Abstract
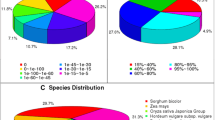
Accelerated root formation is necessary for successful cuttage in a number of woody plants, such as mulberry (Morus alba L.). However, little information regarding the molecular mechanisms involved in this type of root formation is available. Here, we compared transcriptome changes during different root formation periods (0, 4, and 8 days) by conducting high-throughput sequencing to understand the molecular mechanisms of root formation in mulberry softwood cuttings. Approximately 88.2 million, 88.2 million, and 52.8 million transcriptome sequencing reads were obtained for the three time periods, 0, 4, and 8 days, respectively. These reads were then assembled into 61,687 (0 day), 61,651 (4 days), and 59,457 (8 days) contigs by utilizing the de novo method, and a large number of differentially expressed genes were identified at various developmental stages. These assembled contigs were annotated using Kyoto Encyclopedia of Genes and Genomes (KEGG), as well as the simple sequence repeat (SSR). Mulberry had a complex response to adventitious rooting. Genes that might be associated with mulberry stem cutting root formation were also identified here. Furthermore, the reliability of the sequencing results was validated by performing quantitative real-time polymerase chain reaction (qRT-PCR). These results provide a comprehensive background of the molecular biology of mulberry development, in particular regarding its stem cutting root formation development.




Similar content being viewed by others
References
Asano K, Vornlocher HP, Richter-Cook NJ, Merrick WC, Hinnebusch AG, Hershey JW (1997) Structure of cDNAs encoding human eukaryotic initiation factor 3 subunits. Possible roles in RNA binding and macromolecular assembly. J Biol Chem 272(43):27042–27052
Beltran-Pena E, Ortiz-Lopez A, Sanchez de Jimenez E (1995) Synthesis of ribosomal proteins from stored mRNAs early in seed germination. Plant Mol Biol 28(2):327–336
Brinker M, van Zyl L, Liu W, Craig D, Sederoff RR, Clapham DH, Arnold S (2004) Microarray analyses of gene expression during adventitious root development in Pinus contorta. Plant Physiol 135(3):1526–1539
Cheng J, Xiao L, Wang P (2010) A kind of stereo seedling method and products (Application number: 200710132106). China Patent
De Hoff PL, Brill LM, Hirsch AM (2009) Plant lectins: the ties that bind in root symbiosis and plant defense. Mol Genet Genomics 282(1):1–15
De Klerk GJ, Hanecakova J (2008) Ethylene and rooting of mung bean cuttings. The role of auxin induced ethylene synthesis and phase-dependent effects. Plant Growth Regul 56(2):203–209
Dewitte W, Murray JA (2003) The plant cell cycle. Annu Rev Plant Biol 54(1):235–264
Du W (2010) Study on lenticels-derived rooting mechanism in mulberry softwood cuttings by artificial induction. Sericultural Research Institute, Jiangsu University of Science and Technology
Feng J, Qi L, Sun X (2010) Construction of suppression subtractive hybridization library and expression pattern of a few genes involved in rooting of Larix cuttings. Sci Silvae Sin 46:27–34
Forsyth C, Staden JV (1987) Cytokinin metabolism in tomato plants. I. metabolites of zeatin in decapitated roots. J Plant Physiol 128(1):1–10
Gaedeke N, Klein M, Kolukisaoglu U et al (2001) The Arabidopsis thaliana ABC transporter AtMRP5 controls root development and stomata movement. EMBO J 20(8):1875–1887
Gibson SI (2005) Control of plant development and gene expression by sugar signaling. Curr Opin Plant Biol 8(1):93–102
Haissig BE (1972) Meristematic activity during adventitious root primordium development: influences of endogenous auxin and applied gibberellic acid. Plant Physiol 49(6):886–892
Himanen K, Vuylsteke M, Vanneste S, Vercruysse S, Boucheron E, Alard P, Chriqui D, Van Montagu M, Inze D, Beeckman T (2004) Transcript profiling of early lateral root initiation. Proc Natl Acad Sci U S A 101(14):5146–5151
Ho S, Chao Y, Tong W, Yu S (2001) Sugar coordinately and differentially regulates growth- and stress-related gene expression via a complex signal transduction network and multiple control mechanisms. Plant Physiol 125:877–890
Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32(suppl 1):D277–D280
Koch KE (1996) Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol 47(1):509–540
Li M, Leung DWM (2000) Starch accumulation is associated with adventitious root formation in hypocotyl cuttings of Pinus radiata. J Plant Growth Regul 19(4):423–428
Li L, Peng J, Bai R (2010) Analysis of the genetic relationships in Chinese Ziziphus with SRAP markers. Agric Sci China 9(9):1278–1284
Liu M, Sun L, Zhang X, Nie H (2011) Physiological and biochemical analysis of mulberry hardwood cuttings rooting process. China Seric 32(1):9–14
Lu J, Mao Y, Shen L, Peng S, Liu M (2005) Application of AFLP markers for identification of hybrids from open pollinated Dongzao. Acta Hortic Sin 32(4):680–684
Majer C, Xu C, Berendzen KW, Hochholdinger F (2012) Molecular interactions of rootless concerning crown and seminal roots, a LOB domain protein regulating shoot-borne root initiation in maize (Zea mays L.). Philos Trans R Soc Lond B Biol Sci 367(1595):1542–1551
Murphy A, Peer WA, Taiz L (2000) Regulation of auxin transport by aminopeptidases and endogenous flavonoids. Planta 211(3):315–324
Pan G, Lou CF (2008) Isolation of an 1-aminocyclopropane-1-carboxylate oxidase gene from mulberry (Morus alba L.) and analysis of the function of this gene in plant development and stresses response. J Plant Physiol 165(11):1204–1213
Perrot-Rechenmann C (2010) Cellular responses to auxin: division versus expansion. CSH Perspect Biol 2(5):a001446
Popescu SC, Tumer NE (2004) Silencing of ribosomal protein L3 genes in N. tabacum reveals coordinate expression and significant alterations in plant growth, development and ribosome biogenesis. Plant J 39(1):29–44
Potato Genome Sequencing Consortium (2011) Genome sequence and analysis of the tuber crop potato. Nature 475(7355):189–195
Qi H, Xue X, Chen Y (2011) Effects of ethylene on root induction of netted melon (Cucumis melo L.) in vitro. J Shenyang Agric Univ 42(1):26–30
Rabijns A, Barre A, Van Damme EJ, Peumans WJ, De Ranter CJ, Rougé P (2005) Structural analysis of the jacalin-related lectin MornigaM from the black mulberry (Morus nigra) in complex with mannose. FEBS J 272(14):3725–3732
Rouhier N, Couturier J, Jacquot JP (2006) Genome-wide analysis of plant glutaredoxin systems. J Exp Bot 57(8):1685–1696
Saez-Vasquez J, Gallois P, Delseny M (2000) Accumulation and nuclear targeting of BnC24, a Brassica napus ribosomal protein corresponding to a mRNA accumulating in response to cold treatment. Plant Sci 156(1):35–46
Sharkey TD, Yeh S, Wiberley AE, Falbel TG, Gong D, Fernandez DE (2005) Evolution of the isoprene biosynthetic pathway in kudzu. Plant Physiol 137(2):700–712
Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446(7136):640–645
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10(1):57–63
Compliance with Ethical Standards
We have known the ethical responsibilities of authors and declare to comply with ethical standards. We have reviewed the final version of the manuscript and approved it for publication. To the best of our knowledge and belief, this manuscript has not been published in whole or in part nor is it being considered for publication elsewhere.
Conflict of Interest
The authors declare that they have no competing interests.
Human and animal studies
This article does not contain any studies with human or animal subjects.
Funding
This work was supported by China Agriculture Research System (Grant No. CARS-22).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Du, W., Ban, Y., Nie, H. et al. A Comparative Transcriptome Analysis Leads to New Insights into the Molecular Events Governing Root Formation in Mulberry Softwood Cuttings. Plant Mol Biol Rep 34, 365–373 (2016). https://doi.org/10.1007/s11105-015-0927-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-015-0927-1