Abstract
Systemin is a signal peptide that promotes the response to wounding and herbivore attack in tomato. This 18-amino acid peptide is released from a larger precursor, prosystemin. To study the role of systemin as a modulator of defense signaling, we generated tomato (Solanum lycopersicum) transgenic plants that overexpress the prosystemin cDNA. We carried out a transcriptomic analysis comparing two different transgenic events with the untransformed control. The Gene Ontology categories of the 503 differentially expressed genes indicated that several biological functions were affected. Systemin promotes the expression of an array of defense genes that are dependent on different signaling pathways and it downregulates genes connected with carbon fixation and carbohydrate metabolism. These alterations present a degree of overlap with the response programs that are classically associated to pathogen defense or abiotic stress protection, implying that end products of the systemin signaling pathway may be more diverse than expected. We show also that the observed transcriptional modifications have a relevant functional outcome, since transgenic lines were more resistant against very different biotic stressors such as aphids (Macrosiphum euphorbiae), phytopathogenic fungi (Botrytis cinerea and Alternaria alternata) and phytophagous larvae (Spodoptera littoralis). Our work demonstrated that in tomato the modulation of a single gene is sufficient to provide a wide resistance against stress by boosting endogenous defense pathways. Overall, the data provided evidence that the systemin peptide might serve as DAMP signal in tomato, acting as a broad indicator of tissue integrity.
Similar content being viewed by others
Introduction
Plants have evolved a variety of defense mechanisms against biotic stress, including chemical compounds (e.g., toxins and anti-nutrient proteins), physical barriers (e.g., waxes, thorns, and trichomes), and indirect defenses that entail the recruitment of predators and parasitoids (Wu and Baldwin 2010). Many defenses are inducible, activated following the perception of herbivores or pathogens (Kessler and Baldwin 2002). In plants, a composite signaling cascade transcriptionally controls the production of a variety of chemically diverse metabolites involved in plant defense. Many induced responses are mediated by the interplay of relatively few phytohormones (Pieterse et al. 2012). There is a general agreement that jasmonic acid (JA), salicylic acid (SA), and ethylene (Et) play a dominant role in defense in nearly all plant species (Pieterse et al. 2012; Robert-Seilaniantz et al. 2011).
Insects, as well as other agents of biotic stress, induce also a defense reaction in undamaged distal tissues (systemically) (Heil and Bostock 2002; Kessler and Baldwin 2002; Wu and Baldwin 2010). This response implies the presence of signals that move through vascular tissues. Systemic signals can be different but are also related to jasmonates and salicylates (Farmer and Ryan 1990; Holopainen and Blande 2012). Moreover, volatile hormones (e.g., ethylene) and their derivatives (e.g., methyl jasmonate and methyl salicylate) participate in the systemic response to insect herbivores (Farmer and Ryan 1990; Holopainen and Blande 2012).
Systemin was the first identified plant bioactive peptide (Pearce et al. 1991). It was isolated from tomato as a potent inducer of protease inhibitors in both local and systemic leaves (Pearce et al. 1991). This 18 amino acids (aa) peptide is released from the C-terminal region of a larger precursor of 200 aa, called prosystemin (Mcgurl et al. 1992). Genetic evidences indicated that systemin most likely generates and amplifies a systemic signal, supporting the JA pathway (Lee and Howe 2003; Li et al. 2002; Schilmiller and Howe 2005). Upon wounding, systemin initiates a cascade that leads to the induction of early signaling components of the defense pathway and the resulting accumulation of molecules directly and indirectly affecting pests (Corrado et al. 2007; Mcgurl et al. 1994; Ryan 2000). At the cellular level, systemin induces a depolarization of the plasma membrane, the induction of mitogen-activated protein kinases (MAPKs), an increase of intracellular Ca2+ concentration and the activation of a phospholipase A2 (PLA2) (Ryan 2000; Sun et al. 2011). The latter is involved in the release of linolenic acid from membrane phospholipids, which primes the octadecanoid pathway and the downstream biosynthesis of 12-oxophytodienoic acid (OPDA) and jasmonic acid (Ryan 2000; Sun et al. 2011).
The role of systemin in the wounding or herbivore response was elucidated in a number of studies exploiting gain- or loss-of-function mutants. The overexpression of prosystemin induced the accumulation of protease inhibitors (PIs) that degrade essential amino acids in the herbivore midgut (Chen et al. 2005; Mcgurl et al. 1994). The silencing of prosystemin gene strongly reduces the plant response to herbivores (Orozcocardenas et al. 1993). More recently, it has been demonstrated that systemin plays a wider and more complex role. Tomato plants overexpressing prosystemin produced more volatile organic compounds (VOCs) (Corrado et al. 2007; Degenhardt et al. 2010) and were more attractive for parasitoids (Corrado et al. 2007). Furthermore, systemin is involved in tomato resistance against necrotrophic phytopathogens (Diaz et al. 2002; El Oirdi et al. 2011). Prosystemin overexpression also increased resistance to saline stress (Orsini et al. 2010).
All these features are usually discussed taking into account the overlap between different stress-related pathways. It has been demonstrated that the SA- and JA-pathways represent a flexible signaling network in plants (Koornneef and Pieterse 2008; Kunkel and Brooks 2002; Mur et al. 2006), which allows to fine tune the response against invaders and to minimize the fitness and metabolic cost associated to the defense reaction (Robert-Seilaniantz et al. 2011). As the different signaling pathways involved in stress response rely on modules composed of negative and positive regulatory components (Robert-Seilaniantz et al. 2011), it is likely that their balanced crosstalk is important to achieve a specific plant response to biotic stress. This is also demonstrated by the fact that the modification of the hormonal crosstalk in attacked plants seems to be a common strategy for different biotic stress (El Oirdi et al. 2011; Robert-Seilaniantz et al. 2011; Walling 2000).
Recent progress has revealed that biotic stress can generate larger than expected sets of differentially expressed genes, which usually include a significant proportion of sequences that do not code for proteins directly involved in plant defense (Bilgin et al. 2010; Coppola et al. 2013; De Vos et al. 2005; Kerchev et al. 2012; Thompson and Goggin 2006). These works also indicated that the interactions between signaling pathways are complex and not yet fully discovered. However, whether or not connected transmission processes that originate from a single signaling molecule may cooperate to influence significantly various pathological outcomes has not been frequently addressed.
Signaling peptides like systemin are believed to be crucial in the control of important plant functions by modulating different response pathways. With the aim of understating the molecular mechanisms that trigger the tomato response to phytophagous pests, we analyzed the transcriptomic changes associated to the overexpression of prosystemin and evaluated its effect against different damaging organisms. Our data led to a more comprehensive understanding of the systemin defense network, highlighted that the positive and negative regulation of the expression of genes involved in diverse molecular pathways has an extensive functional outcome in the tomato-biotic stress interactions, and indicated that the systemin peptide has the ability to reinforce responses towards different biotic stresses.
Materials and Methods
Tomato Genetic Transformation
Considering that the hybrid tomato cultivar “Better Boy” that was originally transformed with a construct overexpressing prosystemin cDNA (Mcgurl et al. 1994) is a VFN variety (i.e., resistant to Verticillium wilt, Fusarium wilt, root-knot nematodes and aphids), we used as a recipient a susceptible genotype, the “Red Setter” variety, more appropriate to study systemin effect on biotic stresses. Furthermore, since this variety is a non-hybrid cultivar, it is also possible to have genetically uniform progenies. Seeds of Solanum lycopersicum L. “Red Setter” were sterilized and germinated in vitro on TRI1 medium (2.2 g l−1 MS salts, 0.2 mg l−1 thiamine, 50 mg l−1 myo-Inositol, 0.2 mg l−1 IAA, 15 g l−1 sucrose; pH 5.9) solidified with 8 g l−1 agar (Duchefa). Agrobacterium tumefaciens (strain C5851) containing the pMZ plasmid (Rocco et al. 2008) was grown in AB medium (60 g l−1 K2HPO4, 20 g l−1 NaH2PO4, 20 g l−1 NH4Cl, 3 g l−1 KCl, 5 g l−1 glucose, 6 g l−1 MgSO4·7H2O, 0.2 g l−1 CaCl2, 50 mg l−1 FeSO4·7H2O) supplemented with 500 mg l−1 streptomycin and 50 mg l−1 kanamycin. A. tumefaciens-mediated transformation of tomato cotyledons was performed as described (Vanroekel et al. 1993) with a different co-cultivation time (2 days) and without using a feeder layer. Putative transformants were selected on kanamycin (50 mg l−1) and transferred to sterile soil in an environmental chamber 26 + 2 °C with a photoperiod of 16:8 h light/dark. Homozygous T2 populations from two independent single-copy transgenic events were selected for molecular analysis and bioassays.
Molecular Analysis of Plants
DNA was isolated from leaves of 2-week-old plants as described (Fulton et al. 1995). For Southern blot analysis, total DNA (5 μg) was digested with Hind III (Promega), resolved into a 0.7 % (w/v) agarose gel and transferred to a Hybond-N membrane (Amersham) and cross-linked to the membrane by UV (120 mJ) as reported (Corrado et al. 2005). A DIG-labeled probe was obtained using Sys Fw and Sys Rv primers (Supplementary Table 1). Filters were pre-hybridized, incubated with 500 ng of a DIG-labeled probe and washed according to the instruction of the DIG-High Prime DNA Labeling and Detection Starter Kit (Roche). Probe-target hybrids were detected with an enzyme-linked chemiluminescent immunoassay using the CDP-Star substrate (Roche).
The isolation of total RNA from leaves of 4-week-old plants, the synthesis of the first strand cDNA and real-time PCRs were performed as already reported (Corrado et al. 2012). Gene expression analysis was carried out using two technical replicates for each of the three biological replicates per samples. Relative quantification of gene expression was carried out using the 2−ΔΔCt method (Livak and Schmittgen 2001). The statistical significance was evaluated using the Student’s t test. The housekeeping gene EF-1α was used as an endogenous reference gene for the normalization of the expression levels of the target genes. Primers and their main features are reported in the Supplementary Table 1.
For western blot analysis, total soluble proteins were isolated from leaves. Briefly, 0.5 g of leaf tissue were finely grounded in liquid nitrogen and suspended in 300 μl of extraction buffer (6 M Urea, 50 mM Tris–HCl pH 7.5, 50 mM NaCl, 5 mM EDTA; pH 8.0). Proteins were resolved by SDS-PAGE using a 6 % (w/v) stacking-12 % (w/v) separation polyacrylamide gel in a Mini-Protein II (Bio-Rad) apparatus, and transferred onto nitrocellulose membrane by electroblotting with a Mini Trans-Blot Cell system (Bio-Rad). The membrane was probed with anti-prosystemin polyclonal antibody (Narváez-Vásquez and Ryan 2004) (dilution 1:1000), and anti-rabbit IgGs conjugated with peroxidase (Santa Cruz Biotechnology; dilution 1:2500) as described (Tortiglione et al. 2003). Detected proteins were visualized using a chemiluminescent detection system (ECL, GE Healthcare) using Hyperfilm ECL (GE Healthcare). Molecular weights were estimated by comparison with the PageRuler Plus Prestained Protein Ladder (Fermentas).
Two-Color Microarray-Based Gene Expression Analysis
We used three biological replicates per genotype. For each replicate, three leaves of a 4-week-old plant were pooled to reduce noise arising from biological variation. Leaf tissue was powdered in liquid nitrogen and homogenized in Qiazol (Qiagen). Total RNA was extracted using Plant RNeasy mini kit (Qiagen) according to the manufacturer’s protocol. Samples were analyzed with the 2100 Bioanalyzer system (Agilent Technologies) for sizing, quantitation, and quality control of RNA. Only samples with a 260/280 nm absorbance >1.8 and a 260/230 nm absorbance >2 were amplified in the presence of cyanine-3- or cyanine-5-labeled CTP using the Agilent Low Input Quick Amp Labeling kit (Agilent Technologies). Samples were purified using the RNeasy mini spin columns (Qiagen). The quality of labeled targets was determined by calculating the amount of cDNA produced, the picomoles of dye incorporated and the frequency of incorporation, with a NanoDrop 1000 (Thermo Scientific). Equal amounts of cRNAs (825 ng) from control and a transgenic line were mixed together and hybridized to the Tomato Gene Expression Microarray 4x44K (Agilent Technologies) at 65 °C for 17 h in an Agilent Hybridization Oven (G2545A) at 10 rpm. Slides were washed with the Gene Expression Wash buffer 1 for 1 min at room temperature, the Gene Expression Wash buffer 2 for 1 min at 37 °C, and treated with the Stabilization and Drying Solution for 30 s at room temperature. Slides were scanned with a dual-laser microarray scanner (G2565AA, Agilent Technologies) and image data were processed using the Feature Extraction v. 10 software (Agilent Technologies). Raw data and associated sample information were processed by Genespring GX 10 (Agilent Technologies). Statistical analysis was performed using background-corrected mean signal intensities from each dye channel. Microarray data were normalized using intensity-dependent global normalization (LOWESS). Differentially expressed RNAs were identified using a filtering by the Benjamini and Hochberg False Discovery Rate (p < 0.05) and a minimum of a two-fold variation in expression compared to untransformed controls.
Functional Annotation
The sequence of each differentially expressed probe was downloaded from NCBI starting from the available information provided by the microarray manufacturer (Agilent Technologies). Functional annotation was carried out by sequence analysis using the Blast2GO software (Gotz et al. 2008). Briefly, a BlastX similarity search against the nr NCBI protein database was performed to retrieve a maximum of 20 homologous hits per query. GO-term mapping and annotation were retrieved using NCBI as well as non-redundant reference protein database (PSD, UniProt, Swiss-Prot, TrEMBL, RefSeq, GenPept, and PDB Full Gene Ontology DB). Additional annotations (e.g., the recovery of implicit “Biological Process” and “Cellular Component” GO terms from “Molecular Function” annotations) were implemented using ANNEX. Completion of the functional annotation with protein domain information was obtained with InterProScan 5.0. Mapping of enzymatic activities into molecular pathways was acquired from the KEGG database.
Lepidoptera Bioassay
Spodoptera littoralis larvae were grown in an environmental chamber at 25 ± 2 °C, 70 ± 5 % RH and fed with an artificial diet composed by 41.4 g l−1 wheat germ, 59.2 g l−1 brewer’s yeast and 165 g l−1 corn meal, supplemented with 5.9 g l−1 ascorbic acid, 1.8 g l−1 methyl 4-hydroxybenzoate and 29.6 g l−1 agar. About 60 eggs were hatched on this artificial diet and allowed to grow until the second instar. Uniform second instar larvae, were selected and separated in three groups of 12–15 members and each group was used to evaluate larval weight and survival rate after feeding on control, RSYS 24 and RSYS32 leaf disks. Single larvae were isolated in a tray well (Bio-Ba-8, Color-Dec, Italy) covered by perforated plastic lids (Bio-Cv-1, Color-Dec Italy), containing 2 % agar (w/v) to create a moist environment required to keep turgid the experimental tomato leaf disks. Larvae were daily offered leaf disks of uniform size, initially of 2 cm2, later of 3, 4, and 5 cm2 following larvae growth.
Plastic trays were kept at 28 °C 16:8 h light/dark photoperiod. Larval weight and mortality were recorded until pupae development. Fourth instar larvae were transferred into plastic boxes containing vermiculite for pupae development. Data were collected from two experimental replications.
Necrotrophic Fungi Bioassay
Five-week-old plants were tested for resistance to Botrytis cinerea and Alternaria solani as previously described (Corrado et al. 2005). Briefly, spores of B. cinerea and A. solani were suspended in sterile distilled water, filtered through sterile Kimwipes (Kimberly-Clark) to remove fragments of hyphae and adjusted to a concentration of 1 · 106 conidia per ml. Ten μl of the spore suspension were applied between the leaf veins, using four different inoculation points per leaf. The assay with detached leaves was carried out using four plants per genotype. For each plant, two leaves were placed on sponges soaked in sterile water and incubated in a growth chamber at 23 °C, under 16:8 h light/dark photoperiod and 90 % RH. The size of the lesions was measured after 48, 72, and 96 h. For the whole plant assay, inocula were performed on four plants per genotype, and lesions were measured after 48 and 96 h. Lesion dimensions were measured using a digital caliber (Neiko 01407A).
Aphid Bioassays
A clonal culture of Macrosiphum euphorbiae was reared on S. lycopersicum cv. “San Marzano” in an environmental chamber at 20 ± 2 °C, 65 ± 5 % RH and a 16:8 h light/dark photoperiod. For bioassays, 4-week-old plants were placed inside wood frame cages covered with mesh and infested with synchronized 1-day-old nymphs of M. euphorbiae. Assays were carried out at 20 ± 2 °C, 65 ± 5 % RH, 16:8 h light/dark photoperiod using in total 34 Red Setter and 52 RSYS24 plants. Aphids were free to move on the same plant or to reject the host by dropping off. As new-born nymphs are very sessile during their first days of life (Klingauf 1987), the number of nymphs present on each plant after 24 h was used as an estimate of adult preference (Poch et al. 1998). Mortality was calculated counting the number of surviving aphids after 48 h. Values were normalized using the Henderson-Tilton formula (Dent 2000). Subsequently, the number of aphids and the presence of exuviae were monitored daily for 10 days.
For the weight increase assay, 20 adult apterous aphids were placed on three plants per genotype. Aphids were weighted before being transferred to the host plant and after 48 h of feeding. The statistical significance of the weight increase was assessed by a Student’s t test. The weight of the aphids was not statistically different between experimental groups at the beginning of the test (p < 0.01).
Results
Generation and Analysis of the RSYS Transgenic Lines
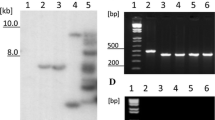
Tomato plants (S. lycopersicum L.) were stably transformed via Agrobacterium with a construct containing prosystemin cDNA under the control of the Cauliflower Mosaic Virus 35S RNA promoter. A schematic representation of the transgenic T-DNA is shown in Fig. 1a. Putative transformants, named RSYS, were screened by PCR (not shown) and Southern blot hybridization to confirm the presence and the number of T-DNA insertions (Fig. 1b). Transgene expression was analyzed by using a real-time RT-PCR approach. Figure 1c reports the quantification of prosystemin expression relative to the untransformed control. The accumulation of the prosystemin pro-hormone in leaves was monitored by Western blot assay (Fig. 1d). A protein with an apparent molecular weight of around 37 kDa was detected in the transgenic lines. The apparent mass of the prosystemin protein does not correspond to the predicted mass (23 kDa), most likely because of the high percentage (44 %) of charged amino acids (Delano et al. 1999). Among the 13 transformants that overexpressed prosystemin cDNA, two lines, namely RSYS24 and RSYS32, were selected, as they show a high level of expression and have a single transgenic locus. The phenotype of these transgenic lines is presented in the Supplementary Figure 1. As prosystemin overexpression generates a signal that constitutively induces proteinase inhibitors synthesis (Mcgurl et al. 1994), we also evaluated the expression level of three genes coding for proteinase inhibitors (proteinase inhibitor I, II and metallocarboxypeptidase proteinase inhibitor) by real-time RT-PCR. The three genes were upregulated in the transgenic plants (Fig. 1e), indicating the generation of a signal that induces constitutive proteinase inhibitor synthesis.
Functional Annotation of the Genes Activated by Prosystemin Overexpression
The transcriptomic changes imposed by the constitutive prosystemin overexpression were monitored by using the Tomato Gene Expression 4x44k array (Agilent). A comparative gene expression analysis was performed with cDNAs from leaves of the two selected transgenic genotypes (RSYS24 and RSYS32) and Red Setter untransformed controls. After filtering, differentially expressed transcripts were identified using a fold-change cut-off ≥2.0 and a p < 0.05 (Benjamini and Hochberg False Discovery Rate). This analysis identified 689 differentially expressed probes that, according to the available reference tomato genome, correspond to 503 genes. Approximately, three quarters (74 %) were upregulated in the transgenic lines, while the remaining downregulated (Supplementary Tables 2 and 3, respectively). To validate microarray results, the expression of ten differentially expressed genes was analyzed by real-time PCR. The Supplementary Figure 2 shows the concordance between microarray log two-fold change and log two real-time RQ values, on a linear scale. The results indicated a significant (p < 0.01) and high correlation (0.96; Pearson coefficient) between the two data sets.
Functional annotation of the differentially expressed sequences and data mining on the resulting annotations was based predominantly on the gene ontology (GO) vocabulary. For the “biological process” domain, a broader overview of the ontology content was achieved by using the GO-plant slim list, to limit the detail of the specific fine-grained terms.
Considering the “biological process” GO terms, the multilevel distribution of the upregulated sequences indicated that “response to stress”, “cellular amino acid metabolic process”, and “catabolic process” were the most relevant series of molecular events or functions affected by prosystemin overexpression (Fig. 2a; Supplementary Table 4). In quantitative terms, other relevant processes were “response to biotic stimulus”, “secondary metabolic process”, “transcription, DNA-dependent”, and “transport”. For the downregulated sequences, the most represented GO terms were “catabolic process”, “response to stress”, “carbohydrate metabolic process”, and “transport” (Fig. 2b; Supplementary Table 5).
The categorization of the annotated genes using the KEGG database indicated differences between the metabolic pathways affected by the up- and the downregulated genes (Supplementary Tables 6 and 7, respectively). According to the absolute number of sequences involved, prosystemin mainly increased the metabolism of several amino acids (cysteine, methionin, arginine, proline, tyrosine, glycine, serine, and trypthophan), of the alpha-linolenic and fatty-acid pathways, and of phenylpropanoids-related pathways. Prosystemin downregulated genes whose enzymatic activities were often included in carbohydrate metabolism, such as starch, sucrose, galactose, fructose, mannose, amino sugar, and nucleotide sugar (i.e., involved in glycosylation) metabolism, as well as carbon fixation.
Effects of Prosystemin Overexpression on the Tomato Transcriptome
As expected, prosystemin increased the expression of genes involved in the biosynthesis of jasmonates (i.e., a chloroplastic 13S-lipoxigenase, a peroxisomal ketoacyl-thiolase, a phospholipase and an allele oxide synthase) and of a mono-oxygenase involved in the production of linoleic acid. Nonetheless, the Jasmonate Zim Domain (JAZ) protein 1 and 3 were also overexpressed. In Arabidopsis, JAZ proteins are considered negative regulators for JA-responsive genes (Chung et al. 2008). Degradation of JAZ repressors triggered by ubiquitin-proteasome induces the expression of transcription factors such as MYC2 driving the expression of JA-related genes in response to tissue injury or other stresses. Systemin-dependent activation of the tomato JAZ proteins 1 and 3 is most likely a consequence of the high level of the JA signaling, so it could represent a strategy of feedback control or energetic costs reduction. Besides their roles in regulating developmental processes, plant hormones are involved in signaling networks related to the stress response (Robert-Seilaniantz et al. 2011). Moreover, the transgenic lines overexpressed ethylene transcription factors (6) and two genes coding for amino acid hydrolases that release active IAA from conjugates. Although plausible, it is not experimentally known if these tomato enzymes also hydrolyze amino acid-conjugated forms of JA such as the jasmonoyl isoleucine, considered the bioactive JA derivative needed for long-distance signaling (Katsir et al. 2008). Prosystemin also upregulated a gene coding for a det2-like (steroid 5-alpha-reductase-like) protein and of a 3-beta-hydroxysteroid-delta-isomerase-like, enzymes involved in the early steps of brassinosteroid biosynthesis (Fujioka et al. 1997). Furthermore, SA responsive genes, such as those coding for different classes of pathogenesis-related proteins (i.e., osmotin, β-glucosidase 13, pathogenesis-related protein 10, pto-responsive gene 1 protein, defensin, endochitinase, and thaumatin) were upregulated. The data imply that different hormone signal transductions play a role in systemin-mediated responses. Accordingly, the effect of prosystemin overexpression involved also molecular sensors whose abundance is affected by a range of environmental cues such as Ca2+ and reactive oxygen species (ROS) (Kessler and Baldwin 2002; Wu and Baldwin 2010). Calcium-dependent kinases and calmodulins were overexpressed in the transgenic plants, as well as antioxidant enzymes (four glutathione S-transferase-like proteins; a thioredoxin m).
Twelve overexpressed sequences are connected with enzymatic activities associated to the phenylpropanoid pathway. In plants, the biosynthesis of all phenylpropanoids begins with the amino acids phenylalanine and tyrosine. In the transgenic lines, both phenylalanine ammonia-lyases (PAL), responsible for the transformation of l-phenylalanine into trans-cinnamic acid, and trans-cinnamate 4-monooxygenases/hydrolases (C4H), responsible for the transformation of trans-cinnamate into 4-hydroxycinnamate (p-coumaric acid), were overexpressed. Phenylpropanoid metabolism generates an array of secondary metabolites, including phenylpropanoid esters, flavonoids, and anthocyanins, which have been linked to plant signaling and defense against biotic or abiotic stress (Naoumkina et al. 2010; Vogt 2010). Moreover, the products derived from the dehydrogenative polymerization of the three monolignol precursors deriving from the PP are also essential for the biosynthesis of lignin. The biosynthetic flux of monolignols is controlled in different steps but, in addition to the phenylalanine supply, the elements that are considered important are the cinnamate 4-hydroxylase (C4H) and of p-coumarate 3-hydroxylase (CH3) activities (Anterola and Lewis 2002; Naoumkina et al. 2010). Prosystemin increased also the expression of a p-coumarate 3-hydroxylase, suggesting a variation of the strength and stiffness of the secondary cell wall. In addition, genes related to the flavonoid biosynthesis (e.g., leucoanthocyanidin dioxygenase-like, 2-oxoglutarate Fe-dependent dioxygenase-like, caffeoyl-o-methyltransferases, hydroxycinnamoyl transferases) were also upregulated in leaves. Besides their role in pigmentation of flowers, fruits pollen, and seeds, flavonoids play a role in plant adaptation to harsh environmental conditions, as well as in the interaction between plants and biotic stress. For instance, the amount and type of flavonoids are important determinants of the leaf taste and can strongly deter feeding by herbivores (Aron and Kennedy 2008; Dixon 2005; Harborne and Williams 2000).
The most abundant class of direct gene products involved in defense was represented by protease inhibitors (18 sequences), such as inhibitors of metallocarboxypeptidases, cysteine, trypsin, and kunitz-type proteases. Other overexpressed genes that can directly affect pest performance were eight polyphenol oxidases, three leucine aminopeptidases, two serine carboxypeptidases, two arginine decarboxylases, and one threonine deaminase. The data indicated that prosystemin can boost the synthesis of a range of proteins to impair digestive processes in the insect gut. Moreover, genes coding for proteinase inhibitors showed the highest fold change, representing, for instance, eight of the top ten highly expressed genes (Supplementary Table 2). Polyphenol oxidases represent the second highly expressed class of genes. Prosystemin also activated genes that may affect indirect defense against pests. The transcriptional analysis indicated the upregulation of gene-encoding enzymes of the terpenoid biosynthetic pathway (3-beta-hydroxysteroid-Delta (8), Delta (7)-isomerase-like, geraniol 8-hydroxylase-like, putative monoterpene synthase 1). Compounds produced by this pathway are major components of the Volatile Organic Compound blend (Holopainen and Blande 2012; Walling 2000).
Transgenic plants also overexpressed genes involved in abiotic stress tolerance such as two spermidine- and five spermine-synthases, which promote the formation of higher molecular weight polyamines (Alcazar et al. 2010). Other overexpressed genes were those coding for a desiccation responsive protein, a late embryogenesis-abundant protein, a DNA repair protein uvh3-like, and a annexin. Annexins are calcium-dependent phospholipid-binding proteins with a peroxidase activity, which often participate in plant response to stress (Gorecka et al. 2007).
Prosystemin also alters the expression of genes involved in primary metabolic processes and several genes involved in sugar metabolism and carbon fixation were downregulated. Specifically, seven genes involved in the amino sugar and nucleotide metabolism, seven in starch and sucrose metabolism, five in galactose metabolism, three in fructose and mannose metabolism and three genes in carbon fixation (including a ribulose bisphosphate carboxylase small chain) were expressed at lower level in the transgenic lines. The data imply that such reduction could be a genetically programmed plant response, rather than an effect of a direct interaction with an herbivore (e.g., water loss from damaged tissue), as widely reported (Berger et al. 2007; Ishiga et al. 2009; Nabity et al. 2013). Despite these evidences, recently Attaran and collaborators (2014) demonstrated that in Arabidopsis the downregulation of the expression of genes involved in photosynthesis following defense elicitation is not always correlated with the reduction of photosynthetic efficiency. The authors suggest that, in Arabidopsis, the photosynthetic system endures variation in the expression of components without significant reduction in quantum efficiency of photosystem II. Therefore, we cannot exclude that, although RSYS plants show a significant downregulation of transcripts of photosynthesis associated genes, their photosynthetic efficiency remain unaltered.
Transcriptional control of stress-responsive genes is a crucial part of the plant response and systemin also affected the expression of transcription factors (TF). In addition to the above-mentioned ERF genes, a member of the other main families of TFs related to stress (i.e., bZIP and WRKY proteins) were differentially expressed. Among others, TFs related to heat-stress were differentially expressed (two up- and one downregulated). A MYC-type ICE1-like TF that increases the amount of different osmolyte (Feng et al. 2013) was overexpressed, while a BIM1-like TF, a bHLH protein involved in brassinosteroid signaling, was downregulated.
Evaluation of the Resistance Against Phytophagous Larvae
In order to assess in our transgenic lines the resistance conferred by prosystemin overexpression against Lepidoptera (McGurl et al. 1992), plants were assayed against the cotton leafworm (S. littoralis), a pest on vegetables, fruits, flowers, and other crops. After 8 days, the weight of the larvae fed with transgenic leaves started to be significantly lower (Fig. 3a). Larvae survival rate was also reduced during the whole bioassay (Fig. 3b). After 25 days of feeding, the survival rate was 24 % for RSYS24 and 12 % for RSYS32, compared to 84 % for the untransformed plants. The data indicated that prosystemin overexpression compromised severely both growth and survival of the S. littoralis larvae.
Molecular analysis of the transgenic lines. a T-DNA map of the pMZ binary vector used for plant genetic transformation showing the restriction site used for Southern blot analysis (not to scale). Arrows represent promoter sequences, and black triangles indicate the location of the primers P1 (Sys Fw) and P2 (Sys Rv) used for probe synthesis and RT-PCR. A dashed line underlines the region hybridized in the Southern analysis. LB T-DNA left border sequence; 35S CaMV35S RNA gene promoter; prosystemin prosystemin cDNA sequence; rbcS poly(A) addition sequence of the pea rbcS; nptII neomycin phosphotransferase II coding sequence; nos nopaline synthase promoter; RB T-DNA right border sequence. b Southern blot analysis. 1 Red Setter, 2 RSYS 24, 3 RSYS 32. Numbers at the left margin indicate marker fragment sizes in kilobase pairs. c Relative quantification (RQ) of the prosystemin expression by real-time RT-PCR. 1 Red Setter, 2 RSYS 24, 3 RSYS 32. Quantities (RQ) are shown relative to the calibrator genotype Red Setter. Data statistical significance was calculated using Student’s t test (*p < 0.05; **p < 0.01). d Western blot analysis. 1 Red Setter, 2 RSYS 24, 3 RSYS 32. Numbers at the left margin indicate marker sizes in kilodaltons. e Relative quantification (RQ) of the expression of proteinase inhibitors by real-time RT-PCR. Pin I proteinase inhibitor I, Pin II proteinase inhibitor II, MCPI metallocarboxypeptidase proteinase inhibitors. Quantities (RQ) are shown relative to the calibrator genotype Red Setter. Data statistical significance was calculated using Student’s t test (*p < 0.05; **p < 0.01)
Multilevel pie charts representation of GO-annotation results. Relative distribution of “Biological Process” terms following GO classification of the upregulated (a) and downregulated (b) genes (sequence cut-off > 5). A multilevel pie was obtained using the lowest GO terms per branch that fulfill the annotation weight criteria (sequence abundance; cut-off > 5)
Effect of prosystemin overexpression on Spodoptera littoralis larvae. a Average weight (±s.d.) of S. littoralis larvae feed on transgenic or control leaves. At each time points, values were significantly different between controls and transgenic lines starting from 8 days (*p < 0.05; Student’s t test); b Survival rate of larvae feed on transgenic or control leaves
Prosystemin Overexpression Increases Tomato Resistance Against Necrotrophic Fungi
We observed that among overexpressed genes in RSYS plants there are transcripts induced by necrotrophic fungi and therefore possibly involved in the enhancement of inducible defenses. In addition, systemin was previously reported to be involved in plant tolerance to B. cinerea (Diaz et al. 2002; El Oirdi et al. 2011). Therefore, we evaluated the performance of 4-week-old RSYS plants to necrotrophic fungi. Two different experiments were carried out: one on whole plants and the other on detached leaves. Disease severity was quantified by measuring the necrotic areas. The results of the first experiment are shown in Fig. 4. Transgenic leaves of both RSYS24 and RSYS32 plants display a strong reduction of fungi induced lesions (Fig. 4). We then extended the evaluation of tolerance to A. solani, a necrotrophic pathogen known to attack tomato leaves. In this experiment, we used detached leaves of RSYS24 that were inoculated with A. solani or B. cinerea (Fig. 5). For both pathogens, lesions development was significantly reduced in prosystemin overexpressing leaves.
Effect of prosystemin overexpression on Botrytis cinerea in whole plant assay. Response to B. cinerea artificial infection by leaves from untransformed (Red Setter) and transformed lines (RSYS 24 and RSYS 32). The graphs display the average (±s.d.) of the lesion size at 48, 96 h post infestation (hpi). At each time points, values were significantly different between controls and transgenic lines starting (*p < 0.05; Student’s t test)
Effect of prosystemin overexpression on Alternaria solani and Botrytis cinerea in detached leaf assay. Response to A. solani and B. cinerea artificial infection bt untransformed (Red Setter) and transformed leaves (RSYS 24). The graphs display the average (±s.d.) of the lesion size at 48, 72, and 96 h post infestation (hpi). At each time points, values were significantly different between controls and transgenic lines starting (*p < 0.05; Student’s t test)
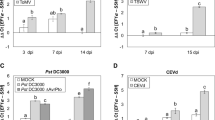
The resistance to necrotrophic fungi has been described to be also JA/Et-mediated (AbuQamar et al. 2008; Glazebrook 2005). Therefore, to get new insights on the role of prosystemin as enhancer of the resistance to fungal pathogens, we examined the expression levels of genes that are studied often in relation to plant-microbe interaction. From the differentially expressed genes, we evaluated the expression of osmotin, extension, and miraculin in leaves of Red Setter plants following B. cinerea infection. Furthermore, we also studied the expression of the Arginase 2. Arginases are enzymes involved in the biosynthesis of polyamines that accumulate in plants in response to environmental stimuli (Chen et al. 2004). B. cinerea infestation of ‘Red Setter’ plants upregulates the selected prosystemin-activated genes, implying that the resistance of the transgenic plants is likely to be dependent by the constitutive expression of an array of different genes (Fig. 6).
Expression analysis of selected genes in Red Setter leaves following Botrytis cinerea inoculation. Relative quantification (log RQ) of gene expression by real-time RT-PCR of selected differentially expressed genes. The 2−ΔΔCt values were significantly different between treated and control plants (*p < 0.05; **p < 0.01; Student’s t test)
Prosystemin Overexpression Increases Aphid Resistance in Tomato
To evaluate if the constitutive expression of prosystemin increases resistance against phloem-feeders, plants of the RSYS 24 line were assayed against M. euphorbiae. The results of the no-choice test experiments indicated that M. euphorbiae did not avoid transgenic plants compared to the untransformed control (Table 1). On the other hand, both longevity and mortality were different between the genotypes under investigation. An increased tolerance of the transgenic plants was also indicated by the lower adults weight increase. Although aphids tend to colonize the transgenic plants when no other choice is offered, the data indicated that prosystemin overexpression enhances aphid antibiosis in tomato.
The possible overlap between the pathways activated by prosystemin and by the aphid feeding was investigated analyzing the expression level, following aphid infestation, of genes selected from the microarray study. Leaves were harvested 96 h post infestation, when aphids have established a feeding site. The data indicated that the selected genes (Pin I, Pin II, KPI, LAP, TD, PR1, and WRKY40) were differentially expressed following aphid infestation (Fig. 7). An interesting difference between the response to aphids and to prosystemin was related to PR1 expression, a protein typically produced in plants in the event of a pathogen attack. PR1 was downregulated in the RSYS plant but induced by aphid feeding. The latter is, for instance, consistent with data in Arabidopsis thaliana seedlings (Moran and Thompson 2001) and indicated that tomato response to aphids is complex and shares components with both fungal pathogens and herbivores (Coppola et al. 2013; Smith and Boyko 2007; Thompson and Goggin 2006).
Expression analysis of selected genes in Red Setter leaves following aphid infestation. Relative quantification (RQ) of gene expression by real-time RT-PCR of selected differentially expressed genes. Pin I proteinase inhibitor I, Pin II proteinase inhibitor II, KPI Kunitz-type proteinase inhibitor family protein, LAP leucine amino peptidase, TD threonine deaminase, PR1 pathogenesis-related protein 1. The 2−ΔΔCt values were significantly different between treated and control plants (*p < 0.05; **p < 0.01; Student’s t test)
Discussion
The molecular response to wounding and/or herbivory has been investigated in different plant species. Considerable progress has been gained from model systems but different species can also show remarkable differences in relation to JA and systemin signaling (Li et al. 2004; Schmidt and Baldwin 2006). Systemin-mediated defense has represented an important model to elucidate plant signal transduction in response to insect attack in tomato. Following the seminal work of Ryan’s group (Mcgurl et al. 1994), one of the highly expressing transgenic lines has been used in a number of studies to shed light on defense mechanisms against chewing insects and mechanical wounding (Bergey et al. 1999; Chen et al. 2005; Constabel et al. 1995; Dombrowski et al. 1999; Jacinto et al. 1997; Li et al. 2003). Currently, it is widely accepted that the multicomponent response to pests requires an extensive genetic reprogramming and metabolic re-allocation. In absence of a stress, defenses are kept under genetic control in order to be activated only when plants sense the danger. An advantage of the overexpression of prosystemin is to activate JA responses with high specificity without the plausible complicating effects of tissue damage or attacker-derived molecules that target other physiological processes in the host (e.g., water loss, tissue removal, etc.). However, the impact of the systemin peptide on tomato transcriptome, as well as its possible functional outcome in other interactions, has not been systematically addressed. Moreover, large-scale microarray studies on plant response to pests are relatively scarce in tomato and other cultivated Solanaceae (Coppola et al. 2013; Lawrence et al. 2008; Scranton et al. 2013; Strassner et al. 2002; Uppalapati et al. 2005).
The expression analysis, based on two independent single-copy transgenic lines, revealed a higher number of systemin-associated genes than previously reported (Bergey et al. 1999; Ryan 2000), implying a broader effect of this peptide in tomato. The activation of the oxylipin pathway and related defense compounds represents a substantial effect especially considering the fold-change of the gene expression, because genes coding for different proteins that could directly affect pests were the most highly expressed. The inhibitory spectrum of upregulated proteinase inhibitors was wide, as they include members acting against serine, cysteine, and metalloproteases as well as proteins involved in the catabolism of essential amino acids in the insects gut. Despite the metabolic cost of producing a range of proteins with similar biological function, a concerted expression of different PI is essential to provide a long-term influence on phytophagous pests (Lawrence and Koundal 2002; Zhu-Salzman et al. 2008). Nonetheless, prosystemin effects were not restricted to the JA-pathway, since several genes that are dependent on different signaling cascades and pathways were differentially regulated. The expression profile of the transgenic lines presented a degree of overlap with the response programs that are classically associated to pathogen defense or abiotic stress protection. Moreover, different stress-hormone related genes were differentially expressed, providing further evidence that multiple hormone pathways are integrated for defense and wound responses (Robert-Seilaniantz et al. 2011). On the other hand, the transcriptional analysis revealed also a degree of specificity in systemin-induced responses, as subsets of genes related to the JA, SA, ethylene, and brassinosteroid pathways were differentially expressed. As a part of the defense response, the majority of the downregulated genes were connected to carbon metabolism and fixation (Bilgin et al. 2010). As this effect was present in absence of a direct interaction with a biotic stress, out data provided support to the hypothesis that downregulation of photosynthesis is a plant-driven response to the perception of stress, rather than a secondary physiological response to tissue damage and/or water loss. Another relevant feature of prosystemin accumulation was related to phenolic secondary metabolites. Given the wide role of these compounds, only a metabolic analysis would provide sufficient detail to infer a specific effect. Nonetheless, plant phenols and polyphenols are known to increase resistance against fungal pathogens and phytophagous insects. Moreover, if present, an increased lignin biosynthesis and cell wall cross-linking activity would play a broad role in defense against different biotic stressors (Bhuiyan et al. 2009; Nicholson and Hammerschmidt 1992). In addition, taking also into account the previously reported increase in salt-tolerance of prosystemin overexpressing plants (Orsini et al. 2010), the transcriptomics analysis also suggests that prosystemin has the potential to improve both abiotic and biotic stress tolerance of tomato.
Considering the genes differentially expressed, we evaluated the possible functional outcome of prosystemin constitutive overexpression. The bioassay indicated that transgenic plants were more resistant to Lepidoptera as expected, but also to aphids and necrotrophic fungi. While the latter can be discussed considering the JA-mediated defenses against necrotrophic pathogens, our data also indicated that other systemin-activated genes, which are not directly linked to the JA-pathway, should contribute to the enhanced performance of the transgenic plants. Genes coding for component of the basal immunity to pathogens (osmotin, miraculin, and extensin) were overexpressed. In tomato, a possible effect of prosystemin on the activation of pathogen defense genes has not yet been investigated but a similar effect was described in potato on different pathogen-associated genes (Bhattacharya et al. 2013). Similarly, although tomato response to aphids and to systemin overproduction is not identical in respect to JA- and SA-dependent genes, our data demonstrated that systemin-response is effective against M. euphorbiae. We did not observe differences in the host-acceptance, implying increased plant toxicity. The change in the quality of the host plant is congruent with the inducible production of components directly affecting host feeding. Exogenous JA treatment reduces aphid population growth on a susceptible tomato cultivar (Cooper et al. 2004; Cooper and Goggin 2005) as well as in other species (Gao et al. 2007). More recently, it has been also shown that antisense suppression of the FAD7 gene in tomato, which is associated with enhanced levels of SA, reduces aphid infestations (Avila et al. 2012). Either SA- or JA-dependent acquired resistance in tomato may have a direct negative effect on phloem-feeding insects, as already proposed (Cooper et al. 2004). Considering that the transcriptional analysis indicated that prosystemin overexpression activates genes associated not only to the octadecanoid pathway, further studies are needed to determine how the possible variation in SA, JA, and ET level contributes to the outcome of tomato-biotic stress interaction. Overall, the GO categories of the differentially expressed genes indicated that systemin-dependent plant response is based on the link between the regulation of photosynthesis, hormonal signaling, and production of compounds involved in direct and indirect defense against stress (Kerchev et al. 2012; Wu and Baldwin 2010). The transcriptional response is associated with upregulation of some primary metabolic pathways that would support the accumulation of a number of defense proteins, without a compensatory enhancement of carbon metabolism and photosynthesis. We previously reported that the overexpression of prosystemin associated to a lower CO2 assimilation rate and intracellular concentration (Corrado et al. 2011). The enhanced re-routing of metabolism towards primary and secondary compounds involved in defense represents a cost that would explain the slower growth of the plants (Corrado et al. 2011).
Even though the gene ontology of the differentially expressed genes cross biotic-abiotic stress boundaries, these effects are consistent with the proposed role of systemin as enhancer of the herbivore response. Herbivores cause not only tissue removal and cell damage but also increase water loss and susceptibility to opportunistic pathogens.
Besides systemin, a number of plant signaling peptides that activate genes counteracting herbivores or pathogens invasions have been isolated and characterized in different species (Huffaker et al. 2006; Narvaez-Vasquez et al. 2007; Pearce and Ryan 2003; Ren and Lu 2006). In the plant kingdom, defense signaling peptides seem to be involved more often in plant-pathogen interactions (Albert 2013), leading to the proposition that tomato and other members of the Solanoideae subfamily (i.e., potato, nightshade and pepper) may have adapted systemin to strengthen the inducible response to herbivores (Constabel et al. 1998). While genes and signals produced by herbivore or pathogen attack are considered different, the components of the transduction pathways and related gene products may have evolved with some degree of conservation of structure, function or both. It has also been reported that B. cinerea enhances prosystemin expression in tomato (El Oirdi et al. 2011). The transcriptomic changes and the broad resistance against biotic stress that we observed suggest that systemin is a component of multiple defense pathways, supporting the proposition that plant-derived peptide elicitors have a more general role in amplifying defense signaling pathways against herbivores, fungal pathogens, or more likely both (Albert 2013; Pearce et al. 2008). Systemin peptide should be then considered a damage-associated molecular patterns (DAMPs) molecule, which is released by stressed cells to dictate an innate immune reaction (Albert 2013; Howe and Jander 2008; Yamaguchi and Huffaker 2011). Moreover, our study implies also that, at least in the absence of a direct interaction with a biotic stressor, a clear dichotomy between pathogen- and herbivore-specific defense pathways does not always exist (Felton and Korth 2000), consistent with the proposed presence in tomato of reciprocal, plant-mediated positive interactions (Stout et al. 1999).
In conclusion, our work provides a detailed overview of the transcriptomic modifications determined by prosystemin overexpression in tomato, leading to a more comprehensive understanding of systemin defense signaling network. The broad effect of prosystemin suggests that a vast array of plant defense processes may be controlled by a relatively reduced number of endogenous molecules. Finally, our data demonstrate that tomato resistance against different biotic stresses can be significantly enhanced by promoting the expression of a single gene that encodes a key component of the inducible defensive systemic signaling system. The effect of prosystemin might well influence different aspects of plant-environment interaction, for instance, not limited to biotic stressors but also affecting beneficial organisms or neighboring plants. Future studies are needed to increase our understanding on how the modulation of an endogenous signal can influence tomato performance in a more complex scenario.
References
AbuQamar S, Chai MF, Luo HL, Song FM, Mengiste T (2008) Tomato protein kinase 1b mediates signaling of plant responses to necrotrophic fungi and insect herbivory. Plant Cell 20:1964–1983
Albert M (2013) Peptides as triggers of plant defence. J Exp Bot 64:5269–5279
Alcazar R, Altabella T, Marco F, Bortolotti C, Reymond M, Knocz C, Carrasco P, Tiburcio AF (2010) Polyamines: molecules with regulatory functions in plant abiotic stress tolerance. Planta 231:1237–1249
Anterola AM, Lewis NG (2002) Trends in lignin modification: a comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity. Phytochemistry 61:221–294
Aron PM, Kennedy JA (2008) Flavan-3-ols: nature, occurrence and biological activity. Mol Nutr Food Res 52:79–104
Attaran E, Major IT, Cruz JA, Rosa BA, Koo AJ, Chen J, Kramer DM, He SY, Howe GA (2014) Temporal dynamics of growth and photosynthesis suppression in response to jasmonate signaling. Plant Physiol 165(3):1302–1314
Avila CA, Arévalo-Soliz LM, Jia L, Navarre DA, Chen Z, Howe GA, Meng QW, Smith JE, Goggin FL (2012) Loss of function of fatty acid desaturase7 in tomato enhances basal aphid resistance in a salicylate-dependent manner. Plant Physiol 158:2028–2041
Berger S, Benediktyová Z, Matouš K, Bonfig K, Mueller MJ, Nedbal L, Roitsch T (2007) Visualization of dynamics of plant-pathogen interaction by novel combination of chlorophyll fluorescence imaging and statistical analysis: differential effects of virulent and avirulent strains of P. syringae and of oxylipins on A. thaliana. J Exp Bot 58:797–806
Bergey DR, Orozco-Cardenas M, de Moura DS, Ryan CA (1999) A wound- and systemin-inducible polygalacturonase in tomato leaves. Proc Natl Acad Sci U S A 96:1756–1760
Bhattacharya R, Koramutla MK, Negi M, Pearce G, Ryan CA (2013) Hydroxyproline-rich glycopeptide signals in potato elicit signalling associated with defense against insects and pathogens. Plant Sci 207:88–97
Bhuiyan NH, Selvaraj G, Wei Y, King J (2009) Role of lignification in plant defense. Plant Signal Behav 4:158–159
Bilgin DD, Zavala JA, Zhu J, Clough SJ, Ort DR, DeLucia EH (2010) Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ 33:1597–1613
Chen H, McCaig BC, Melotto M, He SY, Howe GA (2004) Regulation of plant arginase by wounding, jasmonate, and the phytotoxin coronatine. J Biol Chem 279:45998–46007
Chen H, Wilkerson CG, Kuchar JA, Phinney BS, Howe GA (2005) Jasmonate-inducible plant enzymes degrade essential amino acids in the herbivore midgut. Proc Natl Acad Sci U S A 102:19237–19242. doi:10.1073/pnas.0509026102
Chung HS, Koo AJ, Gao X, Jayanty S, Thines B, Jones AD, Howe GA (2008) Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol 146(3):952–64
Constabel CP, Bergey DR, Ryan CA (1995) Systemin activates synthesis of wound-inducible tomato leaf polyphenol oxidase via the octadecanoid defense signaling pathway. Proc Natl Acad Sci U S A 92:407–411
Constabel CP, Yip L, Ryan CA (1998) Prosystemin from potato, black nightshade, and bell pepper: primary structure and biological activity of predicted systemin polypeptides. Plant Mol Biol 36:55–62
Cooper WR, Goggin FL (2005) Effects of jasmonate-induced defenses in tomato on the potato aphid, Macrosiphum euphorbiae. Entomol Exp Appl 115:107–115
Cooper WC, Jia L, Goggin FL (2004) Acquired and R-gene-mediated resistance against the potato aphid in tomato. J Chem Ecol 30:2527–2542
Coppola V, Coppola M, Rocco M, Digilio MC, D’Ambrosio C, Renzone G, Martinelli R, Scaloni A, Pennacchio F, Rao R, Corrado G (2013) Transcriptomic and proteomic analysis of a compatible tomato-aphid interaction reveals a predominant salicylic acid-dependent plant response. BMC Genomics 14 doi:10.1186/1471-2164-14-515
Corrado G, Bovi PD, Ciliento R, Gaudio L, Di Maro A, Aceto S, Lorito M, Rao R (2005) Inducible expression of a Phytolacca heterotepala ribosome-inactivating protein leads to enhanced resistance against major fungal pathogens in tobacco. Phytopathology 95:206–215
Corrado G, Sasso R, Pasquariello M, Iodice L, Carretta A, Cascone P, Ariati L, Digilio MC, Guerrieri E, Rao R (2007) Systemin regulates both systemic and volatile signaling in tomato plants. J Chem Ecol 33:669–681
Corrado G, Agrelli D, Rocco M, Basile B, Marra M, Rao R (2011) Systemin-inducible defence against pests is costly in tomato. Biol Plant 55:305–311
Corrado G, Alagna F, Rocco M, Renzone G, Varricchio P, Coppola V, Coppola M, Garonna A, Baldoni L, Scaloni A, Rao R (2012) Molecular interactions between the olive and the fruit fly Bactrocera oleae. BMC Plant Biol 12
De Vos M et al (2005) Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant Microbe Interact 18:923–937
Degenhardt DC, Refi-Hind S, Stratmann JW, Lincoln DE (2010) Systemin and jasmonic acid regulate constitutive and herbivore-induced systemic volatile emissions in tomato, Solanum lycopersicum. Phytochemistry 71:2024–2037
Delano JP, Dombrowski JE, Ryan CA (1999) The expression of tomato prosystemin in Escherichia coli: a structural challenge. Protein Expr Purif 17:74–82
Dent D (2000) Insect pest management, 2nd edn. CABI, Wellingford
Diaz J, ten Have A, van Kan JAL (2002) The role of ethylene and wound signaling in resistance of tomato to Botrytis cinerea. Plant Physiol 129:1341–1351
Dixon RA (2005) Engineering of plant natural product pathways. Curr Opin Plant Biol 8:329–336
Dombrowski JE, Pearce G, Ryan CA (1999) Proteinase inhibitor-inducing activity of the prohormone prosystemin resides exclusively in the C-terminal systemin domain. Proc Natl Acad Sci U S A 96:12947–12952
El Oirdi M, El Rahman TA, Rigano L, El Hadrami A, Rodriguez MC, Daayf F, Vojnov A, Bouarab K (2011) Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. Plant Cell 23:2405–2421
Farmer EE, Ryan CA (1990) Interplant communication—airborne methyl jasmonate induces synthesis of proteinase-inhibitors in plant-leaves. Proc Natl Acad Sci U S A 87:7713–7716
Felton GW, Korth KL (2000) Trade-offs between pathogen and herbivore resistance. Curr Opin Plant Biol 3:309–314
Feng HL, Ma NN, Meng X, Zhang S, Wang JR, Chai S, Meng QW (2013) A novel tomato MYC-type ICE1-like transcription factor, SlICE1a, confers cold, osmotic and salt tolerance in transgenic tobacco. Plant Physiol Biochem 73:309–320
Fujioka S, Li J, Choi YH, Seto H, Takatsuto S, Noguchi T, Watanabe T, Kuriyama H, Yokota T, Chory J, Sakurai A (1997) The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell 9:1951–1962
Fulton TM, Chunwongse J, Tanksley SD (1995) Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol Biol Rep 13:207–209
Gao LL, Anderson JP, Klingler JP, Nair RM, Edwards OR, Singh KB (2007) Involvement of the octadecanoid pathway in bluegreen aphid resistance in Medicago truncatula. Mol Plant Microbe Interact 20:82–93
Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43:205–227
Gorecka KM, Thouverey C, Buchet R, Pikula S (2007) Potential role of annexin AnnAt1 from Arabidopsis thaliana in pH-mediated cellular response to environmental stimuli. Plant Cell Physiol 48:792–803
Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36:3420–3435
Harborne JB, Williams CA (2000) Advances in flavonoid research since 1992. Phytochemistry 55:481–504
Heil M, Bostock RM (2002) Induced systemic resistance (ISR) against pathogens in the context of induced plant defences. Ann Bot-Lond 89:503–512
Holopainen JK, Blande JD (2012) Molecular plant volatile communication. In: López-Larrea C (ed) Sensing in nature. Advances in experimental medicine and biology. Springer, 739, pp 17–31
Howe GA, Jander G (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59:41–66
Huffaker A, Pearce G, Ryan CA (2006) An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci U S A 103:10098–10103
Ishiga Y, Uppalapati SR, Ishiga T, Elavarthi S, Martin B, Bender CL (2009) The phytotoxin coronatine induces light-dependent reactive oxygen species in tomato seedlings. New Phytol 181:147–160
Jacinto T, McGurl B, Franceschi V, DelanoFreier J, Ryan CA (1997) Tomato prosystemin promoter confers wound-inducible, vascular bundle-specific expression of the beta-glucuronidase gene in transgenic tomato plants. Planta 203:406–412
Katsir L, Chung HS, Koo AJK, Howe GA (2008) Jasmonate signaling: a conserved mechanism of hormone sensing. Curr Opin Plant Biol 11:428–435
Kerchev PI, Fenton B, Foyer CH, Hancock RD (2012) Plant responses to insect herbivory: interactions between photosynthesis, reactive oxygen species and hormonal signalling pathways. Plant Cell Environ 35:441–453. doi:10.1111/j.1365-3040.2011.02399.x
Kessler A, Baldwin IT (2002) Plant responses to insect herbivory: the emerging molecular analysis. Annu Rev Plant Biol 53:299–328. doi:10.1146/annurev.arplant.53.100301.135207
Klingauf FA (1987) Host plant finding and acceptance. In: Minks AK, Harrewijn P (eds) Aphids: their biology, natural enemies, and control. Elsevier, Amsterdam, pp 209–224
Koornneef A, Pieterse CMJ (2008) Cross talk in defense signaling. Plant Physiol 146:839–844
Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5:325–331
Lawrence PK, Koundal KK (2002) Plant protease inhibitors in control of phytophagous insects. Electron J Biotechnol 5
Lawrence SD, Novak NG, Ju CJT, Cooke JEK (2008) Potato, Solanum tuberosum, defense against Colorado potato beetle, Leptinotarsa decemlineata (Say): microarray gene expression profiling of potato by Colorado potato beetle regurgitant treatment of wounded leaves. J Chem Ecol 34:1013–1025
Lee GI, Howe GA (2003) The tomato mutant spr1 is defective in systemin perception and the production of a systemic wound signal for defense gene expression. Plant J 33:567–576
Li L, Li CY, Lee GI, Howe GA (2002) Distinct roles for jasmonate synthesis and action in the systemic wound response of tomato. Proc Natl Acad Sci U S A 99:6416–6421
Li C, Liu G, Xu C, Lee GI, Bauer P, Ling HQ, Ganal MW, Howe GA (2003) The tomato Suppressor of prosystemin-mediated responses2 gene encodes a fatty acid desaturase required for the biosynthesis of jasmonic acid and the production of a systemic wound signal for defense gene expression. Plant Cell 15:1646–1661
Li L, Zhao Y, McCaig BC, Wingerd BA, Wang J, Whalon ME, Pichersky E, Howe GA (2004) The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16:783–783
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 (Delta Delta Ct) method. Methods 25:402–408
Mcgurl B, Pearce G, Orozcocardenas M, Ryan CA (1992) Structure, expression, and antisense inhibition of the systemin precursor gene. Science 255:1570–1573
Mcgurl B, Orozcocardenas M, Pearce G, Ryan CA (1994) Overexpression of the prosystemin gene in transgenic tomato plants generates a systemic signal that constitutively induces proteinase-inhibitor synthesis. Proc Natl Acad Sci U S A 91:9799–9802
Moran PJ, Thompson GA (2001) Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant Physiol 125:1074–1085
Mur LAJ, Kenton P, Atzorn R, Miersch O, Wasternack C (2006) The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol 140:249–262
Nabity PD, Zavala JA, DeLucia EH (2013) Herbivore induction of jasmonic acid and chemical defences reduce photosynthesis in Nicotiana attenuata. J Exp Bot 64:685–694
Naoumkina MA, Zhao QA, Gallego-Giraldo L, Dai XB, Zhao PX, Dixon RA (2010) Genome-wide analysis of phenylpropanoid defence pathways. Mol Plant Pathol 11:829–846
Narváez-Vásquez J, Ryan CA (2004) The cellular localization of prosystemin: a functional role for phloem parenchyma in systemic wound signaling. Planta 218(3):360–369
Narvaez-Vasquez J, Orozco-Cardenas ML, Ryan CA (2007) Systemic wound signaling in tomato leaves is cooperatively regulated by systemin and hydroxyproline-rich glycopeptide signals. Plant Mol Biol 65:711–718
Nicholson LR, Hammerschmidt R (1992) Phenolic compounds and their role in disease resistance. Annu Rev Phytopathol 30:369–389
Orozcocardenas M, Mcgurl B, Ryan CA (1993) Expression of an antisense prosystemin gene in tomato plants reduces resistance toward Manduca sexta larvae. Proc Natl Acad Sci U S A 90:8273–8276. doi:10.1073/pnas.90.17.8273
Orsini F, Cascone P, De Pascale S, Barbieri G, Corrado G, Rao R, Maggio A (2010) Systemin-dependent salinity tolerance in tomato: evidence of specific convergence of abiotic and biotic stress responses. Physiol Plant 138:10
Pearce G, Ryan CA (2003) Systemic signaling in tomato plants for defense against herbivores—isolation and characterization of three novel defense-signaling glycopeptide hormones coded in a single precursor gene. J Biol Chem 278:30044–30050
Pearce G, Strydom D, Johnson S, Ryan CA (1991) A polypeptide from tomato leaves induces wound-inducible proteinase-inhibitor proteins. Science 253:895–898
Pearce G, Bhattacharya R, Chen YC (2008) Peptide signals for plant defense display a more universal role. Plant Signal Behav 3:1091–1092
Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28:489–521
Poch HLCY, Ponz F, Fereres A (1998) Searching for resistance in Arabidopsis thaliana to the green peach aphid Myzus persicae. Plant Sci 138:209–216
Ren F, Lu YT (2006) Overexpression of tobacco hydroxyproline-rich glycopeptide systemin precursor gene in transgenic tobacco enhances resistance against Helicoverpa armigera larvae. Plant Sci 171:286–292
Robert-Seilaniantz A, Grant M, Jones JDG (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49:317–343. doi:10.1146/annurev-phyto-073009-114447
Rocco M, Corrado G, Arena S, D’Ambrosio C, Tortiglione C, Sellaroli S, Marra M, Rao A, Scaloni A (2008) The expression of tomato prosystemin gene in tobacco plants highly affects host proteomic repertoire. J Proteomics 71:176–185
Ryan CA (2000) The systemin signaling pathway: differential activation of plant defensive genes. BBA-Protein Struct M 1477:112–121
Schilmiller AL, Howe GA (2005) Systemic signaling in the wound response. Curr Opin Plant Biol
Schmidt S, Baldwin IT (2006) Systemin in Solanum nigrum. The tomato-homologous polypeptide does not mediate direct defense responses. Plant Physiol 142:1751–1758
Scranton MA, Fowler JH, Girke T, Walling LL (2013) Microarray analysis of tomato’s early and late wound response reveals new regulatory targets for Leucine aminopeptidase A. PLoS ONE 29314
Smith CM, Boyko EV (2007) The molecular bases of plant resistance and defense responses to aphid feeding: current status. Entomol Exp Appl 122:1–16
Stout AK, Fidantsef AL, Duffeya SS, Bostock RM (1999) Signal interactions in pathogen and insect attack: systemic plant-mediated interactions between pathogens and herbivores of the tomato, Lycopersicon esculentum. Physiol Mol Plant Pathol 54:115–130
Strassner J, Schaller F, Frick UB, Howe GA, Weiler EW, Amrhein N, Macheroux P, Schaller A (2002) Characterization and cDNA-microarray expression analysis of 12-oxophytodienoate reductases reveals differential roles for octadecanoid biosynthesis in the local versus the systemic wound response. Plant J 32:585–601
Sun JQ, Jiang HL, Li CY (2011) Systemin/Jasmonate-mediated systemic defense signaling in tomato. Mol Plant 4:607–615
Thompson GA, Goggin FL (2006) Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects. J Exp Bot 57:755–766
Tortiglione C, Fogliano V, Ferracane R, Fanti P, Pennacchio F, Monti LM, Rao R (2003) An insect peptide engineered into the tomato prosystemin gene is released in transgenic tobacco plants and exerts biological activity. Plant Mol Biol 53:891–902
Uppalapati SR, Ayoubi P, Weng H, Palmer DA, Mitchell RE, Jones W, Bender CL (2005) The phytotoxin coronatine and methyl jasmonate impact multiple phytohormone pathways in tomato. Plant J 42:201–217
Vanroekel JSC, Damm B, Melchers LS, Hoekema A (1993) Factors influencing transformation frequency of tomato (Lycopersicon esculentum). Plant Cell Rep 12:644–647
Vogt T (2010) Phenylpropanoid biosynthesis. Mol Plant 3:2–20
Walling LL (2000) The myriad plant responses to herbivores. J Plant Growth Regul 19:195–216
Wu JQ, Baldwin IT (2010) New insights into plant responses to the attack from insect herbivores. Annu Rev Genet 44:1–24
Yamaguchi Y, Huffaker A (2011) Endogenous peptide elicitors in higher plants. Curr Opin Plant Biol 14:351–357
Zhu-Salzman K, Luthe DS, Felton GW (2008) Arthropod-inducible proteins: broad spectrum defenses against multiple herbivores. Plant Physiol 146:852–858
Acknowledgments
We thank Dr Michelina Ruocco (Istituto per la Protezione delle Piante-CNR, Italy) for providing the fungal pathogens and Prof. A. Schaller (University of Hohenheim, Stuttgart) for the anti-prosystemin IgG. This work was supported by the GenopomPro project (PON R&C 2007–2013) financed by the Italian Ministry of University and Research (MIUR) in cooperation with the European Funds for the Regional Development (FESR), and by the Safe & Smart project (Cluster Agroalimentare Nazionale; CTN01_00230_248064).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1
Phenotype of the transgenic lines RSYS 24 and RSYS 32 four weeks after sowing. (JPEG 414 kb)
Supplementary Figure 2
Microarray validation and concordance with the Real Time results. The graph displays the concordance between log2-microarray fold change and log2-Real Time RQ values on a linear scale (R2=0.93). Each dot represent a gene. The genes analyzed were: wound-induced proteinase inhibitor I (InhI); JA-ZIM domain family protein (JA-ZIM); Kunitz-type proteinase inhibitor family protein (Kunitz); lypoxygenase D (LoxD); Mate efflux family protein (Mate); PPO: polyphenoloxidase (PPO); ProSys: prosystemin; Pathogenesis-related protein 1A1 (PT1); Pto-responsive gene 1 (Pto); SAM: S-Adenosyl Methionine (SAM); Subtilisin: subtilisin-like protease (Subtilisin). (JPEG 176 kb)
Supplementary Table S1
(DOCX 19 kb)
Supplementary Table S2 and S3
(XLSX 61 kb)
Supplementary Table S4 and S5
(XLSX 16 kb)
Supplementary Table S6
(DOCX 14 kb)
Supplementary Table S7
(DOCX 14 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Coppola, M., Corrado, G., Coppola, V. et al. Prosystemin Overexpression in Tomato Enhances Resistance to Different Biotic Stresses by Activating Genes of Multiple Signaling Pathways. Plant Mol Biol Rep 33, 1270–1285 (2015). https://doi.org/10.1007/s11105-014-0834-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-014-0834-x