Abstract
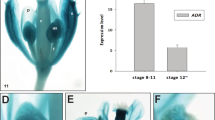
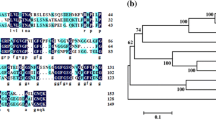
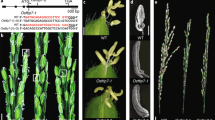
Anther dehiscence, one of the essential steps in pollination and double fertilization, is regulated by a complex signaling pathway encompassing hormones and environmental factors. However, key components underlying the signaling pathway that regulate anther dehiscence remain largely elusive. Here, we isolated a rice mutant anther dehiscence defected 1 (Osadd1) that exhibited defects in anther dehiscence and glume open. Map-based cloning revealed that OsADD1 encoded a GARP (Golden2, ARR-B and Psr1) transcription factor. Sequence analysis showed that a single base deletion in Osadd1 mutant resulted in pre-termination of the GARP domain. OsADD1 was constitutively expressed in various tissues, with more abundance in the panicles. The major genes associated with anther dehiscence were affected in the Osadd1 mutant, and the expression level of the cellulose synthase-like D sub-family 4 (OsCSLD4) was significantly decreased. We demonstrate that OsADD1 regulated the expression of OsCSLD4 by binding to its promoter, and affects rice anther dehiscence.








Similar content being viewed by others
References
Cecchetti V, Brunetti P, Napoli N, Fattorini L, Altamura MM, Costantino P, Cardarelli M (2015) ABCB1 and ABCB19 auxin transporters have synergistic effects on early and late Arabidopsis anther development. J Integr Plant Biol 57:1089–1098
Cecchetti V, Celebrin D, Napoli N, Ghelli R, Brunetti P, Costantino P, Cardarelli M (2017) An auxin maximum in the middle layer controls stamen development and pollen maturation in Arabidopsis. N Phytol 213:1194–1207
Dai M, Zhao Y, Ma Q, Hu Y, Hedden P, Zhang Q, Zhou DX (2007) The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol 144:121–133
Estornell LH, Landberg K, Cierlik I, Sundberg E (2018) SHI/STY genes affect pre- and post-meiotic anther processes in auxin sensing domains in Arabidopsis. Front Plant Sci 9:150
Fujino K, Matsuda Y, Ozawa K, Nishimura T, Koshiba T, Fraaije MW, Sekiguchi H (2008) NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol Genet Genomics 279:499–507
Ghelli R et al (2018) A newly identified flower-specific splice variant of AUXIN RESPONSE FACTOR8 regulates stamen elongation and endothecium lignification in Arabidopsis. Plant Cell 30:620–637
Goldberg RB, Beals TP, Sanders PM (1993) Anther development: basic principles and practical applications. Plant Cell 5:1217
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Hosoda K (2002) Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14:2015–2029
Hu J et al (2010) Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol Biol 73:283–292
Huang T et al (2014) Arabidopsis KANADI1 acts as a transcriptional repressor by interacting with a specific cis-element and regulates auxin biosynthesis, transport, and signaling in opposition to HD-ZIPIII factors. Plant Cell 26:246–262
Ilegems M, Douet V, Meylan-Bettex M, Uyttewaal M, Brand L, Bowman JL, Stieger PA (2010) Interplay of auxin, KANADI and Class III HD-ZIP transcription factors in vascular tissue formation. Development 137:975
Izhaki A, Bowman JL (2007) KANADI and Class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell 19:495
Jibran R, Tahir J, Cooney J, Hunter DA, Dijkwel PP (2017) Arabidopsis AGAMOUS regulates sepal senescence by driving jasmonate production. Front Plant Sci 8:2101
Kang HG, Park S, Matsuoka M, An G (2005) White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C4-type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J 42:901–911
Kanno Y et al (2016) AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat Commun 7:13245
Li M et al (2009) Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J 60:1055–1069
Li L, Shi ZY, Li L, Shen GZ, Wang XQ, An LS, Zhang JL (2010) Overexpression of ACL1 (abaxially curled leaf 1) increased bulliform cells and induced abaxial curling of leaf blades in rice. Mol Plant 3:807–817
Li WQ et al (2017) CLD1/SRL1 modulates leaf rolling by affecting cell wall formation, epidermis integrity and water homeostasis in rice. Plant J 92:904–923
Luan W, Liu Y, Zhang F, Song Y, Wang Z, Peng Y, Sun Z (2011) OsCD1 encodes a putative member of the cellulose synthase-like D sub-family and is essential for rice plant architecture and growth. Plant Biotechnol J 9:513–524
Ma H (2005) Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol 56:393–434
Matsui T, Omasa K (2002) Rice (Oryza sativa L.) cultivars tolerant to high temperature at flowering: anther characteristics. Ann Bot 89:683–687
Matsui T, Omasa K, Horie T (1999) Mechanism of anther dehiscence in rice (Oryza sativa L.). Ann Bot 84:501–506
Mizuno S et al (2007) Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J 50:751–766
Prasad PVV, Boote KJ, Allen LH, Sheehy JE, Thomas JMG (2006) Species, ecotype and cultivar differences in spikelet fertility and harvest index of rice in response to high temperature stress. Field Crops Res 95:398–411
Riechmann JL et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290:2105
Safi A, Medici A, Szponarski W, Ruffel S, Lacombe B, Krouk G (2017) The world according to GARP transcription factors. Curr Opin Plant Biol 39:159–167
Saito H et al (2015) The jasmonate-responsive GTR1 transporter is required for gibberellin-mediated stamen development in Arabidopsis. Nat Commun 6:6095
Sakai H, Aoyama T, Bono H, Oka A (1998) Two-component response regulators from Arabidopsis thaliana contain a putative DNA-binding motif. Plant Cell Physiol 39:1232–1239
Salinas-Grenet H et al (2018) Modulation of auxin levels in pollen grains affects stamen development and anther dehiscence in Arabidopsis. Int J Mol Sci. https://doi.org/10.3390/ijms19092480
Scott RJ, Spielman M, Dickinson HG (2004) Stamen structure and function. Plant Cell 16:S46
Song S, Chen Y, Liu L, See YHB, Mao C, Gan Y, Yu H (2018) OsFTIP7 determines auxin-mediated anther dehiscence in rice. Nat Plants 4:495–504
Steiner-Lange S et al (2003) Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J 34:519–528
Thevenin J et al (2011) The simultaneous repression of CCR and CAD, two enzymes of the lignin biosynthetic pathway, results in sterility and dwarfism in Arabidopsis thaliana. Mol Plant 4:70–82
Wu C, Fu Y, Hu G, Si H, Cheng S, Liu W (2010) Isolation and characterization of a rice mutant with narrow and rolled leaves. Planta 232:313–324
Wu C et al (2012) HRS1 acts as a negative regulator of abscisic acid signaling to promote timely germination of Arabidopsis seeds. PLoS ONE 7:e35764
Xiang JJ, Zhang GH, Qian Q, Xue HW (2012) SEMI-ROLLED LEAF1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol 159:1488–1500
Xu Y et al (2014) Overexpression of OsZHD1, a zinc finger homeodomain class homeobox transcription factor, induces abaxially curled and drooping leaf in rice. Planta 239:803–816
Yang C, Xu Z, Song J, Conner K, Vizcay Barrena G, Wilson ZA (2007) Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19:534–548
Yang C et al (2017) Transcription factor MYB26 is key to spatial specificity in anther secondary thickening formation. Plant Physiol 175:333–350
Yoshikawa T, Eiguchi M, Hibara K, Ito J, Nagato Y (2013) Rice SLENDER LEAF 1 gene encodes cellulose synthase-like D4 and is specifically expressed in M-phase cells to regulate cell proliferation. J Exp Bot 64:2049–2061
Zeng Y, Zhang Y, Xiang J, Uphoff NT, Pan X, Zhu D (2017) Effects of low temperature stress on spikelet-related parameters during anthesis in Indica–Japonica hybrid rice. Front Plant Sci 8:1350
Zhang G-H, Xu Q, Zhu X-D, Qian Q, Xue H-W (2009) SHALLOT-LIKE1 Is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 21:719
Zhao SQ, Hu J, Guo LB, Qian Q, Xue HW (2010) Rice leaf inclination2, a VIN3-like protein, regulates leaf angle through modulating cell division of the collar. Cell Res 20:935–947
Zhao Z et al (2013) A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev Cell 27:113–122
Zhu Q-H, Ramm K, Shivakkumar R, Dennis ES, Upadhyaya NM (2004) The ANTHER INDEHISCENCE1 gene encoding a single MYB domain protein is involved in anther development in rice. Plant Physiol 135:1514
Zou LP et al (2011) Leaf rolling controlled by the homeodomain leucine zipper class IV gene Roc5 in rice. Plant Physiol 156:1589–1602
Acknowledgements
This research was supported by the National Transform Science and Technology Program (2016ZX08001004-002) and National Key Research and Development Program of China (2016YFD0101107, 2016YFD0101801), the Key Laboratory of Biology, Genetics and Breeding of Japonica Rice in the Mid-lower Yangtze River, the Ministry of Agriculture, China, Jiangsu Plant Gene Engineering Research Center, the Jiangsu Collaborative Innovation Center for Modern Crop Production. This study is funded by China Postdoctoral Science Foundation. We are grateful to Professor Wenzhen Liu (China National Rice Research Institute, China) for providing the seeds of Oscsld4/nrl1 mutant.
Author information
Authors and Affiliations
Contributions
Y.J.X., S.M.Y. and Z.G.Z. were involved in conceiving the project, designing the experiments and analyzing the data; W.Y.K., Y.J.X., S.M.Y. and Y.C. were involving in the map-based cloning of the OsADD1 gene; Q.Y.T., W.T.B. and H.Z. were involved in the generation of the transgenic plants; C.L.W. was involved in OsADD1 transient expression in rice protoplasts; Y.J.X. was involved in LUC assay and EMSA; Y.J.X. was involved in writing this article; J.L. and C.M.W. were involved in revising this article; J.L., Z.G.Z. and J.M.W. were involved in data discussions.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xiao, Y., You, S., Kong, W. et al. A GARP transcription factor anther dehiscence defected 1 (OsADD1) regulates rice anther dehiscence. Plant Mol Biol 101, 403–414 (2019). https://doi.org/10.1007/s11103-019-00911-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-019-00911-0