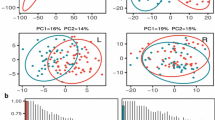
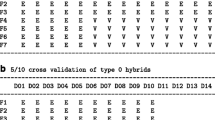
Abstract
Many animal and plant species exhibit increased growth rates, reach larger sizes and, in the cases of crops and farm animals, produce higher yields when bred as hybrids between genetically differing strains, a phenomenon known as hybrid vigour or heterosis. Despite the importance of heterosis, and its extensive genetic analysis, little understanding exists of its molecular basis. We aimed to determine whether characteristics of the leaf transcriptome, as an indicator of the innate functional genetic architecture of a plant line, could be used as markers to predict heterosis and the performance of hybrids, a methodology we term Association Transcriptomics. Relationships between transcript abundance of specific genes and the values of heterosis and heterosis-dependent traits were identified and mathematical models were constructed that relate gene expression characteristics in inbred lines of Arabidopsis thaliana and maize with vegetative biomass and grain yield, respectively, in corresponding hybrids. These models were used to predict, using gene expression data, the performance of additional hybrids. The success of the application in a monocot crop of a methodology developed in a dicot model species indicates that transcriptional markers may have widespread applicability in hybrid breeding.







Similar content being viewed by others
References
Arabidopsis Genome Initiative (2000) Analysis of the genome of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Auger DL, Dogra-Gray A, Ream TS, Kato A, Coe EH Jr, Birchler JA (2005) Nonadditive gene expression in diploid and triploid hybrids of maize. Genetics 169:389–397
Barth S, Busimi AK, Utz HF, Melchinger AE (2003) Heterosis for biomass yield and related traits in five hybrids of Arabidopsis thaliana L. Heynh. Heredity 91:36–42
Bernardo R (1994) Prediction of maize single-cross performance using RFLPs and information from related hybrids. Crop Sci 34:20–25
Bernardo R (1996) Best linear unbiased prediction of maize single-cross performance. Crop Sci 36:50–56
Bernardo R (1998) Predicting the performance of untested single crosses: trait and marker data. In: Lamkey KR, Staub JE (eds) Concepts and breeding of heterosis in crop plants. Crop Science Society of America, Madison, pp 117–127
Bernardo R (1999) Marker-assisted best linear unbiased prediction of singlecross performance. Crop Sci 39:1277–1282
Birchler JA, Bhadra U, Bhadra MP, Auger DL (2001) Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev Biol 234:275–288
Broadley MR, White PJ, Hammond JP, Graham NS, Bowen HC, Emmerson ZF, Fray RG, Iannetta PPM, McNicol JW, May ST (2008) Evidence of neutral transcriptome evolution in plants. New Phytol 180:587–593
Charcosset A, Bonnisseau B, Touchebeuf O, Burstin J, Dubreuil P, Barriere Y, Gallais A, Denis JB (1998) Prediction of maize hybrid silage performance using marker data: comparison of several models for specific combining ability. Crop Sci 38:38–44
Davenport CB (1908) Degeneration, albinism and inbreeding. Science 28:454–455
Duvick DN (1999) Genetic diversity and heterosis. In: Coors CG, Pandey S (eds) Genetics and exploitation of heterosis in crops. American Society of Agronomy, Madison, pp 293–304
East EM (1908) Report of the Connecticut agricultural experiment station for years 1907–1908. New Haven, CT, pp 419–428
Fasoula DA, Fasoula VA (1997) Competitive ability and plant breeding. Plant Breed Rev 14:89–138
Gregory KE, Cundiff LV, Koch RM (1992) Effects of breed and retained heterosis on milk yield and 200-day weight in advanced generations of composite populations of beef cattle. J Anim Sci 70:2366–2372
Guo M, Rupe MA, Danilevskaya ON, Yang X, Hu Z (2003) Genome-wide mRNA profiling reveals heterochronic allelic variation and a new imprinted gene in hybrid maize endosperm. Plant J 36:30–44
Guo M, Rupe MA, Zinselmeier C, Habben J, Bowen BA, Smith OS (2004) Allelic variation of gene expression in maize hybrids. Plant Cell 16:1707–1716
Guo M, Rupe MA, Yang X, Crasta O, Zinselmeie C, Smith OS, Bowen B (2006) Genome-wide transcript analysis of maize hybrids: allelic additive gene expression and yield heterosis. Theor Appl Genet 113:831–845
Hallauer AR (1990) Methods used in developing maize inbreds. Maydica 35:1–16
Hallauer AR, Miranda Filho JB (1988) Quantitative genetics in maize breeding. Iowa State University Press, Ames, Iowa
Hollick JB, Chandler VL (1998) Epigenetic allelic states of a maize transcriptional regulatory locus exhibit overdominant gene action. Genetics 150:891–897
Hua J, Xing Y, Wu W, Xu C, Sun X, Yu S, Zhang Q (2003) Single-locus heterotic effects and dominance by dominance interactions can adequately explain the genetic basis of heterosis in an elite rice hybrid. Proc Natl Acad Sci USA 100:2574–2579
Jordan DR, Tao Y, Godwin ID, Henzell RG, Cooper M, McIntyre CL (2003) Prediction of hybrid performance in grain sorghum using RFLP markers. Theor Appl Genet 106:559–567
Kacser H, Burns JA (1981) The molecular basis of dominance. Genetics 97:639–666
Kosba MA (1978) Heterosis and phenotypic correlations for shank length, body weight and egg production traits in the Alexandria strains and their crosses with Fayoumi chickens. Beitr Trop Landwirtsch Veterinarmed 16:187–198
Liu K, Goodman M, Muse S, Smith JS, Buckler E, Doebley J (2003) Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics 165:2117–2128
Melchinger AE (1999) Genetic diversity and heterosis. In: Coors CG, Pandey S (eds) Genetics and exploitation of heterosis in crops. American Society of Agronomy, Madison, pp 99–1118
Meyer RC, Torjek O, Becher M, Altmann T (2004) Heterosis of biomass production in Arabidopsis. Establishment during early development. Plant Physiol 134:1813–1823
Meyer S, Pospisil H, Scholten S (2007) Heterosis associated gene expression in maize embryos 6 days after fertilization exhibits additive, dominant and overdominant pattern. Plant Mol Biol 63:381–391
Moll RH, Salhuana WS, Robinson HF (1962) Heterosis and genetic diversity in variety crosses of maize. Crop Sci 2:197–198
Moll RH, Lonnquist JH, Velez Fortuno J, Johnson EC (1965) The relationship of heterosis and genetic divergence in maize. Genetics 52:139–144
Nelissen H, Fleury D, Bruno L, Robles P, De Veylder L, Traas J, Micol JL, Van Montagu M, Inzé D, Van Lijsebettens M (2005) The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc Natl Acad Sci USA 102:7754–7759
Ni Z, Sun Q, Liu Z, Wu L, Wang X (2000) Identification of a hybrid-specific expressed gene encoding novel RNA-binding protein in wheat seedling leaves using differential display of mRNA. Mol Gen Genet 263:934–993
O’Neill CM, Gill S, Hobbs D, Morgan C, Bancroft I (2003) Natural variation for seed lipid traits in Arabidopsis thaliana. Phytochemistry 64:1077–1090
Omholt SW, Plahte E, Oyehaug L, Xiang KF (2000) Gene regulatory networks generating the phenomena of additivity, dominance and epistasis. Genetics 155:969–980
Payne RW, Murray DA, Harding SA, Baird DB, Soutar DM (2008) GenStat for Windows (11th Edition). VSN International, Hemel Hempstead, UK
Schrag TA, Mohring J, Maurer HP, Dhillon BS, Melchinger AE, Piepho H-P, Sørensen AP, Frisch M (2009) Molecular marker-based prediction of hybrid performance in maize using unbalanced data from multiple experiments with factorial crosses. Theor Appl Genet 118:741–751
Shull GH (1948) What is “heterosis”? Genetics 33:439–446
Smith OS (1986) Covariance between line per se and testcross performance. Crop Sci 26:540–543
Song R, Messing J (2003) Gene expression of a gene family in maize based on nonlinear haplotypes. Proc Natl Acad Sci USA 100:9055–9060
Stokes D, Morgan C, O’Neill CM, Bancroft I (2007) Evaluating the utility of Arabidopsis thaliana as a model for understanding heterosis in hybrid crops. Euphytica 156:157–171
Stupar RM, Springer NM (2006) Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics 173:2199–2210
Stupar RM, Gardiner JM, Oldre AG, Haun WJ, Chandler VL, Springer NM (2008) Gene expression analyses in maize inbreds and hybrids with varying levels of heterosis. BMC Plant Biol 8:33
Sun QX, Ni ZF, Liu ZY (1999) Differential gene expression between wheat hybrids and their parental inbreds in seedling leaves. Euphytica 106:117–123
Swanson-Wagner RA et al (2006) All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc Natl Acad Sci USA 103:6805–6810
Uzarowska A, Keller B, Piepho H-P, Schwarz G, Ingvardsen C, Wenzel G, Lübberstedt T (2007) Comparative expression profiling in meristems of inbred-hybrid triplets of maize based on morphological investigations of heterosis for plant height. Plant Mol Biol 63:21–34
Vuylsteke M, Kuiper M, Stam P (2000) Chromosomal regions involved in hybrid performance and heterosis: their AFLP (R)-based identification and practical use in prediction models. Heredity 85:208–218
Vuylsteke M, van Eeuwijk F, Van Hummelen P, Kuiper M, Zabeau M (2005) Genetic analysis of variation in gene expression in Arabidopsis thaliana. Genetics 171:1267–1275
Wu LM, Ni ZF, Meng FR, Lin Z, Sun QX (2003) Cloning and characterization of leaf cDNAs that are differentially expressed between wheat hybrids and their parents. Mol Gen Genomics 270:281–286
Xiao JH, Li JM, Yuan LP, Tanksley SD (1995) Dominance is the major genetic-basis of heterosis in rice as revealed by qtl analysis using molecular markers. Genetics 140:745–754
Xiong LZ, Yang GP, Xu CG, Zhang QF, Maroof MAS (1998) Relationships of differential gene expression in leaves with heterosis and heterozygosity in a rice diallel cross. Mol Breed 4:129–136
Zeid M, Schön C-C, Link W (2004) Hybrid performance and AFLP- based genetic similarity in faba bean. Euphytica 139:207–216
Zhu T, Wang X (2000) Large-scale profiling of the Arabidopsis transcriptome. Plant Physiol 124:1472–1476
Acknowledgments
We would like to express out gratitude to Eddie Arthur for his support, advice and encouragement throughout this project. This work was supported by the John Innes Centre Competitive Support Grant, UK Biotechnology and Biological Sciences Research Council PhD studentship 01/A1/G/07546 to D.S., and by Plant Bioscience Limited. GeneChip hybridisations were conducted in the JIC Genome Laboratory by James Hadfield and we are grateful for the help of Gawain Bennett for collation of GeneChip data.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Stokes, D., Fraser, F., Morgan, C. et al. An association transcriptomics approach to the prediction of hybrid performance. Mol Breeding 26, 91–106 (2010). https://doi.org/10.1007/s11032-009-9379-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-009-9379-3