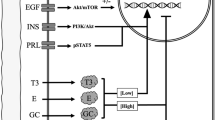
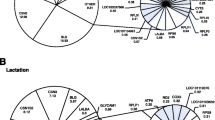
Abstract
The ability to produce and expel milk is important for the health and survival of all mammals. Nevertheless, our understanding of the molecular events underlying the execution of this process remains incomplete. Whilst impaired mammary gland development and lactational competence remains the subject of focused investigations, defects in these events may also be an unintended consequence of genetic manipulation in rodent models. In this technical report, we outline established and emerging methods to characterize lactation phenotypes in genetically-engineered mouse models. We discuss important considerations of common models, optimized conditions for mating and the importance of litter size and standardization. Methods for quantifying milk production and quality, as well as protocols for wholemount preparation, immunohistochemistry and the preparation of RNA and protein lysates are provided. This review is intended to help guide researchers new to the field of mammary gland biology in the systematic analysis of lactation defects and in the preparation of samples for more focused mechanistic investigations.









Similar content being viewed by others
Data Availability
Not applicable. All data generated or analysed during this study are included in this published article.
Abbreviations
- CUBIC:
-
clear unobstructed brain imaging cocktails and computational analysis
- d.p.c.:
-
days post coitum
- E:
-
embryonic day
- HIER:
-
heat-induced epitope retrieval
- L:
-
lactation day
- MG:
-
mammary gland
- MMTV:
-
mouse mammary tumor virus
- NBF:
-
neutral buffered formalin
- PND:
-
post-natal day
- TEB:
-
terminal end bud.
References
Modzelewski AJ, Chen S, Willis BJ, Lloyd KCK, Wood JA, He L. Efficient mouse genome engineering by CRISPR-EZ technology. Nat Protoc. 2018;13:1253–74.
Muñoz-Fuentes V, Cacheiro P, Meehan TF, Aguilar-Pimentel JA, Brown SDM, Flenniken AM, et al. The international mouse phenotyping consortium (IMPC): a functional catalogue of the mammalian genome that informs conservation. Conserv Genet. 2018;19:995–1005.
Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–9.
Palmer CA, Neville MC, Anderson SM, McManaman JL. Analysis of lactation defects in transgenic mice. J Mammary Gland Biol Neoplasia. 2006;11:269–82.
Lloyd-Lewis B, Davis FM, Harris OB, Hitchcock JR, Lourenco FC, Pasche M, et al. Imaging the mammary gland and mammary tumours in 3D: optical tissue clearing and immunofluorescence methods. Breast Cancer Res. 2016;18:127.
Depeters EJ, Hovey RC. Methods for collecting milk from mice. J Mammary Gland Biol Neoplasia. 2009;14:397–400.
Landua JD, Visbal AP, Lewis MT. Methods for preparing fluorescent and neutral red-stained whole mounts of mouse mammary glands. J Mammary Gland Biol Neoplasia. 2009;14:411–5.
Davis B, Fenton SE. Mammary gland. Haschek Rousseaux’s Handb Toxicol Pathol. 3rd ed. 2013. p. 2665–94.
Honvo-Houéto E, Truchet S. Indirect immunofluorescence on frozen sections of mouse mammary gland. J Vis Exp. 2015;106:e53179.
Hens JR, Wysolmerski JJ. Key stages of mammary gland development: molecular mechanisms involved in the formation of the embryonic mammary gland. Breast Cancer Res. 2005;7:220–4.
Cowin P, Wysolmerski J. Molecular mechanisms guiding embryonic mammary gland development. Cold Spring Harb Perspect Biol. 2010;2:a003251.
Stewart TA, Hughes K, Hume DA, Davis FM. Developmental stage-specific distribution of macrophages in mouse mammary gland. Front Cell Dev Biol. 2019;7:250.
Lilja AMM, Rodilla V, Huyghe M, Hannezo E, Landragin C, Renaud O, et al. Clonal analysis of Notch1-expressing cells reveals the existence of unipotent stem cells that retain long-term plasticity in the embryonic mammary gland. Nat Cell Biol. 2018;20:677–87.
Wuidart A, Sifrim A, Fioramonti M, Matsumura S, Brisebarre A, Brown D, et al. Early lineage segregation of multipotent embryonic mammary gland progenitors. Nat Cell Biol. 2018;20:666–76.
Lloyd-Lewis B, Davis FM, Harris OB, Hitchcock JR, Watson CJ. Neutral lineage tracing of proliferative embryonic and adultmammary stem/progenitor cells. Development. 2018;145:164079.
Lloyd-Lewis B, Harris OB, Watson CJ, Davis FM. Mammary stem cells: premise, properties and perspectives. Trends Cell Biol. 2017;8:556–67.
Davis FM, Lloyd-Lewis B, Harris OB, Kozar S, Winton DJ, Muresan L, et al. Single-cell lineage tracing in the mammary gland reveals stochastic clonal dispersion of stem/progenitor cell progeny. Nat Commun. 2016;7:13053.
Van Keymeulen A, Fioramonti M, Centonze A, Bouvencourt G, Achouri Y, Blanpain C. Lineage-restricted mammary stem cells sustain the development, homeostasis, and regeneration of the estrogen receptor positive lineage. Cell Rep. 2017;20:1525–32.
Wang C, Christin JR, Oktay MH, Guo W. Lineage-biased stem cells maintain estrogen-receptor-positive and -negative mouse mammary luminal lineages. Cell Rep. 2017;18:2825–35.
Sreekumar A, Toneff MJ, Toh E, Roarty K, Creighton CJ, Belka GK, et al. WNT-mediated regulation of FOXO1 constitutes a critical axis maintaining pubertal mammary stem cell homeostasis. Dev Cell. 2017;43:436–48.
Scheele C, Hannezo E, Muraro M, Zomer A, Langedijk N, van Oudenaarden A, et al. Identity and dynamics of mammary stem cells during branching morphogenesis. Nature. 2017;542:313–7.
Davis FM. The ins and outs of calcium signalling in lactation and involution: implications for breast cancer treatment. Pharmacol Res. 2016;116:100–4.
Stevenson AJ, Vanwalleghem G, Stewart TA, Condon ND, Lloyd-Lewis B, Marino N, et al. Multiscale imaging of basal cell dynamics in the functionally mature mammary gland. Proc Natl Acad Sci U S A [Internet]. 2020; Available from: https://doi.org/10.1073/pnas.2016905117
Watson CJ, Kreuzaler PA. Remodeling mechanisms of the mammary gland during involution. Int J Dev Biol. 2011;55:757–62.
Prater M, Shehata M, Watson CJ, Stingl J. Enzymatic dissociation, flow cytometric analysis, and culture of normal mouse mammary tissue. Methods MolBiol. 2013;946:395–409.
Smalley MJ. Isolation, culture and analysis of mouse mammary epithelial cells. Methods Mol Biol. 2010;633:139–70.
Smalley MJ, Kendrick H, Sheridan JM, Regan JL, Prater MD, Lindeman GJ, et al. Isolation of mouse mammary epithelial subpopulations: a comparison of leading methods. J Mammary Gland Biol Neoplasia. 2012;17:91–7.
Paine IS, Lewis MT. The terminal end bud: the little engine that could. J Mammary Gland Biol Neoplasia. 2017;22:93–108.
Wagner KU, Wall RJ, St-Onge L, Gruss P, Wynshaw-Boris A, Garrett L, et al. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25:4323–30.
Wagner KU, Ward T, Davis B, Wiseman R, Hennighausen L. Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic Res. 2001;10:545–53.
Rios AC, Fu NY, Lindeman GJ, Visvader JE. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506:322–7.
Ved N, Curran A, Ashcroft FM, Sparrow DB. Tamoxifen administration in pregnant mice can be deleterious to both mother and embryo. Lab Anim. 2019;53:630–3.
Eon JP, Sun X, Nichol P, Saijoh Y, Martin JF, Moon AM, et al. System for tamoxifen-inducible expression of Cre-recombinase from the Foxa2 locus in mice. Dev Dyn. 2008;237:447–53.
Selbert S, Bentley DJ, Melton DW, Rannie D, Lourenço P, Watson CJ, et al. Efficient BLG-Cre mediated gene deletion in the mammary gland. Transgenic Res. 1998;7:387–96.
Chang TH-T, Kunasegaran K, Tarulli GA, De Silva D, Voorhoeve PM, Pietersen AM. New insights into lineage restriction of mammary gland epithelium using parity-identified mammary epithelial cells. Breast Cancer Res. 2014;16:R1.
Stewart TA, Hughes K, Stevenson ASJ, Marino N, Ju AJL, Morehead M, et al. Mammary mechanobiology: mechanically-activated ion channels in lactation and involution. bioRxiv. 2019;649038.
WHITTEN WK. Modification of the oestrous cycle of the mouse by external stimuli associated with the male. J Endocrinol. 1956;13:399–404.
Behringer R, Gertsenstein M, Nagy KV, Nagy A. Selecting female mice in estrus and checking plugs. Cold Spring Harb Protoc. 2016;2016:pdb.prot092387.
Gearhart S, Kalishman J, Melikyan H, Mason C, Kohn DF. Increased incidence of vaginal septum in C57BL/6J mice since 1976. Comp Med. 2004;54:418–21.
Committee. Guide for the Care and Use of Laboratory Animals: Eighth Edition. Guid Care Use Lab Anim. 2011. p. 118.
dos Santos CO, Dolzhenko E, Hodges E, Smith AD, Hannon GJ. An epigenetic memory of pregnancy in the mouse mammary gland. Cell Rep. 2015;11:1102–9.
Lang SLC, Iverson SJ, Bowen WD. Primiparous and multiparous females differ in mammary gland alveolar development: implications for milk production. J Exp Biol. 2012;215:2904–11.
Burkholder T, Foltz C, Karlsson E, Linton CG, Smith JM. Health evaluation of experimental laboratory mice. Curr Protoc Mouse Biol. 2012;2:145–65.
Hurst JL, West RS. Taming anxiety in laboratory mice. Nat Methods. 2010;7:825–6.
Fenton SE, Reiner JL, Nakayama SF, Delinsky AD, Stanko JP, Hines EP, et al. Analysis of PFOA in dosed CD-1 mice. Part 2: disposition of PFOA in tissues and fluids from pregnant and lactating mice and their pups. Reprod Toxicol. 2009;27:365–72.
Reinhardt TA, Lippolis JD, Shull GE, Horst RL. Null mutation in the gene encoding plasma membrane Ca2+-ATPase isoform 2 impairs calcium transport into milk. J Biol Chem. 2004;279:42369–73.
Görs S, Kucia M, Langhammer M, Junghans P, Metges CC. Technical note: Milk composition in mice - methodological aspects and effects of mouse strain and lactation day. J Dairy Sci. 2009;92:632–7.
Schwertfeger KL, McManaman JL, Palmer CA, Neville MC, Anderson SM. Expression of constitutively activated Akt in the mammary gland leads to excess lipid synthesis during pregnancy and lactation. J Lipid Res. 2003;44:1100–12.
Rudolph MC, Wellberg EA, Lewis AS, Terrell KL, Merz AL, Maluf NK, et al. Thyroid hormone responsive protein Spot14 enhances catalysis of fatty acid synthase in lactating mammary epithelium. J Lipid Res. 2014;55:1052–65.
Palmer CA, Lubon H, McManaman JL. Transgenic mice expressing recombinant human protein C exhibit defects in lactation and impaired mammary gland development. Transgenic Res. 2003;12:283–92.
JoVE Science Education Database. Lab animal Reasearch. JoVE: Anesthesia induction and maintenance; 2020.
Davis FM, Janoshazi A, Janardhan KS, Steinckwich N, D’Agostin DM, Petranka JG, et al. Essential role of Orai1 store-operated calcium channels in lactation. Proc Natl Acad Sci. 2015;112:5827–32.
Hughes K, Wickenden JA, Allen JE, Watson CJ. Conditional deletion of Stat3 in mammary epithelium impairs the acute phase response and modulates immune cell numbers during post-lactational regression. J Pathol. 2012;227:106–17.
Radisky DC, Hartmann LC. Mammary involution and breast cancer risk: transgenic models and clinical studies. J Mammary Gland Biol Neoplasia. 2009;14:181–91.
Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–39.
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open source platform for biological image analysis. Nat Methods. 2012;9:676–82.
Sargeant TJ, Lloyd-Lewis B, Resemann HK, Ramos-Montoya A, Skepper J, Watson CJ. Stat3 controls cell death during mammary gland involution by regulating uptake of milk fat globules and lysosomal membrane permeabilization. Nat Cell Biol. 2014;16:1057–68.
Wang Y, Chaffee TS, Larue RS, Huggins DN, Witschen PM, Ibrahim AM, et al. Tissue-resident macrophages promote extracellular matrix homeostasis in the mammary gland stroma of nulliparous mice. Elife. 2020;9:e57438.
Liu Y, Rutlin M, Huang S, Barrick CA, Wang F, Jones KR, et al. Sexually dimorphic BDNF signaling directs sensory innervation of the mammary gland. Science (80- ). 2012;338:1357–60.
Sheridan JM, Visvader JE. Isolation and propagation of mammary epithelial stem and progenitor cells. Methods Mol Biol. 2019. p. 217–29.
Acknowledgements
We thank the UQ Biological Resource Facility staff for assistance with animal husbandry. We also thank animal facilities at the National Institutes of Environmental Health Sciences (NIEHS), University of Cambridge and University of Queensland for their advice and assistance in refining these protocols over the years, as well as Dr. Bethan Lloyd-Lewis (methyl green wholemount protocol) and Dr. Henrike Resemann (IHC protocol).
Funding
This work was supported by the National Health and Medical Research Council of Australia (1141008 and 1138214), the National Stem Cell Foundation of Australia and the Mater Foundation (Equity Trustees / AE Hingeley Trust).
Author information
Authors and Affiliations
Contributions
T.S. and F.D. prepared the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Animal Ethics
All animal work discussed and photographed in this manuscript was carried out in accordance with the Australian Code for the Care and Use of Animals for Scientific Purposes and the Queensland Animal care and Protection Act (2001), with local animal ethics approval. Mice were housed in individually-ventilated cages with food and water available ad libitum and a 12 h light cycle.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Code Availability
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Stewart, T.A., Davis, F.M. Got Milk? Identifying and Characterizing Lactation Defects in Genetically-Engineered Mouse Models. J Mammary Gland Biol Neoplasia 25, 255–272 (2020). https://doi.org/10.1007/s10911-020-09467-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10911-020-09467-y