Abstract
Glycan structures in non-vertebrates are highly variable; it can be assumed that this is a product of evolution and speciation, not that it is just a random event. However, in animals and protists, there is a relatively limited repertoire of around ten monosaccharide building blocks, most of which are neutral in terms of charge. While two monosaccharide types in eukaryotes (hexuronic and sialic acids) are anionic, there are a number of organic or inorganic modifications of glycans such as sulphate, pyruvate, phosphate, phosphorylcholine, phosphoethanolamine and aminoethylphosphonate that also confer a ‘charged’ nature (either anionic or zwitterionic) to glycoconjugate structures. These alter the physicochemical properties of the glycans to which they are attached, change their ionisation when analysing them by mass spectrometry and result in different interactions with protein receptors. Here, we focus on N-glycans carrying anionic and zwitterionic modifications in protists and invertebrates, but make some reference to O-glycans, glycolipids and glycosaminoglycans which also contain such moieties. The conclusion is that ‘charged’ glycoconjugates are a widespread, but easily overlooked, feature of ‘lower’ organisms.
Similar content being viewed by others
Introduction
Glycans modify a range of proteins and lipids, thereby altering their properties and their interactions. All cells have a glycan coat and so the exact nature of glycan structures can determine a wide range of cell-cell, gamete-gamete, host-pathogen, vector-pathogen and host-symbiont interactions; it can be assumed that the glycomic repertoire of a species is formed by evolutionary processes and can define speciation [1]. The basic building blocks of glycans are monosaccharides with hexoses, deoxyhexoses, pentoses and N-acetylhexosamines being very common regardless of whether the glycoconjugate is of animal, plant, protist, fungal or bacterial origin, although the monosaccharide repertoires of bacteria and plants are the most diverse. In addition to these ‘uncharged’ sugars, which can be modified also by moieties such as methyl groups, there are ‘charged’ saccharide and non-saccharide elements. Thereby, hexuronic and sialic acids on glycoconjugates are derived from corresponding uridine diphosphate or cytidine monophosphate donors for the relevant glycosyltransferases; in the case of sulphates, phosphate esters and phosphonates, other activated known or unknown donors are required by the enzymes that perform ‘post-glycosylational modifications’ of glycans.
While sialic and hexuronic acids are well-known in vertebrates as components of glycosaminoglycans, glycoproteins and glycolipids [2,3,4], sulphates are widespread on glycans of many species [5, 6]. Phosphates are known in the context of the mannose-6-phosphate marker for trafficking of lysosomal enzymes [7], but phosphodiesters are less common: exceptions being that phosphoethanolamine is a component of glycosylphosphatidylinositol anchors [8] and ribitol phosphodiesters are part of the modification of dystroglycan [9]. This contrasts with the invertebrates: sialic acids are generally rare, but phosphodiesters and phosphonates have been found on glycoconjugates in a number of species. Hexuronic acids are certainly found in invertebrates, but are less familiar or absent from fungi and protists. In plants, charged modifications (especially hexuronic acids) are on polysaccharides [10, 11], but not on N-glycans.
As N-glycans are the most commonly analysed glycoconjugates and most specific structural information in non-vertebrates is on this class of structure, we will concentrate on these. We also mention the occurrence of anionic and zwitterionic modifications in O-glycan, glycolipid and glycosaminoglycan structures, but exclude glucuronylated and sulphated small metabolites from our discussion. It is probably impossible to be exhaustive in citing all relevant literature and there are other recent reviews which also address neutral glycans in various invertebrates [12,13,14]. We hope readers will appreciate the overall conclusion that hexuronic and sialic acid, sulphate and phospho-based moieties in non-vertebrates can no longer be overlooked, but indeed vastly increase the range of possible glycan structures in a large number of species.
Hexuronic acids
Glucuronic acid is probably the most common hexuronic acid; its activated form (UDP-GlcA) derives from the action of a dehydrogenase on UDP-Glc [15]. In turn, UDP-GlcA can be decarboxylated into UDP-Xyl; thus, any organism capable of making and transferring xylose can also theoretically have glucuronylated glycoconjugates. Indeed, glucuronic acid is known from a variety of glycoconjugates from different species. In terms of mass spectrometric analyses, glucuronic acid has the same mass increment (Δm/z 176 Da) as a methylated hexose. However, hexuronic acids, but not methylhexoses, enable ionisation in both positive and negative MALDI-TOF MS modes; also, aiding definition of these modifications are glucuronidases, which can be used in the analysis of glucuronylated glycans, as well as the enrichment of such structures in the ‘acidic’ fraction upon graphitised carbon-based solid phase extraction [16]. Other known hexuronic acids, include galacturonic acid and iduronic acid, which are respectively components of plant cell wall polysaccharides and animal glycosaminoglycans [17, 18].
We and others have found glucuronic acid residues on N-glycans from molluscs, insects and even a nematode (Fig. 1). In the case of molluscs (specifically one gastropod, Volvarina rubella, and one bivalve, Mytilus edulis), glucuronic acid modifies antennal fucose residues [19, 20]; thus far in insects, whether mosquitoes, the fruit fly, the honeybee or moth species, glucuronic acid was found attached to galactose [21,22,23,24] to form a non-sulphated form of the so-called HNK-1 epitope, while in one filarial nematode (Dirofilaria immitis) N-acetylgalactosamine residues are glucuronylated [25]. Thereby, it is interesting that D. immitis is transmitted via mosquitoes, but it is unknown whether the ‘common’ N-glycan modification with glucuronic acid is relevant to the parasite’s lifecycle. Furthermore, glucuronic acid is the anionic component of acidic glycolipids from flies [26], as part of GlcAβ1,3Galβ1,3GalNAc motifs similar to those found on N-glycan antennae from various insects, whereas GlcA-diacylglycerol is a lipid known from Aspergillus fumigatus [27]. Another yeast pathogen, Cryptococcus neoformans, expresses GlcA side chains linked to galactose residues of a glucuronoxylomannogalactan capsular polysaccharide [28].
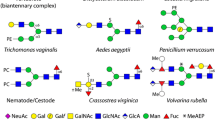
Example glucuronylated, sialylated and sulphated glycans. Various features of N-glycans with anionic moieties are shown to highlight the variations from (i) dipteran (Aedes aegyptii, Anopheles gambiae and Drosophila melanogaster), lepidopteran (Trichoplusia ni and Lymantria dispar) and hymenopteran (Apis mellifera) insect species, (ii) a nematode (specifically Dirofilaria), (iii) the cellular slime mould Dictyostelium discoideum and (iv) Crassostrea virginica (oyster), Volvarina rubella (marine snail; upper depicted arm) and Mytilus edulis (blue mussel; lower depicted arm). Also shown are (v) example ‘mucin-type’ and ‘Notch-type’ O-glycans from nematodes and insects, (vi) glycolipids from Drosophila melanogaster (dipteran; glucuronylated) and Hemicentrotus pulcherrimus (echinoderm; sialylated) and (vii) the glycosaminoglycans (the latter being common to all animals). The glycans are depicted according to the Symbolic Nomenclature for Glycans (see box); MeAEP, methylaminoethylphosphonate; P, phosphate; PC, phosphorylcholine; PE, phosphoethanolamine; PMe, methylphosphate; Pyr, pyruvate, S, sulphate; white circles or boxes indicate undefined hexoses or N-acetylhexosamines. Linkages are defined for proven antennal motifs, while basic trimannosylchitobiosyl cores are assumed for all N-glycans
O-glycans from insects can also be modified with glucuronic acid [22, 29, 30], whether these be of the ‘mucin-type’ (e.g., GlcAβ1,3Galβ1,3GalNAc, Galβ1,3[GlcAβ1,4]GalNAc or ones carrying Hex1HexNAc1HexA1 repeats; the latter found in the lepidopteran HighFive cell line and in mosquitoes) or the ‘Notch-type’ (GlcNAcβ1,3[GlcAβ1,4]Fuc; see Fig. 1); recently, it was shown that O-glycan glucuronylation is important for neuromuscular junction formation in Drosophila [31]. Glucuronic acid is also present on O-glycans from Caenorhabditis elegans [32], while a circulating antigen from the trematode Schistosoma mansoni is a βGalNAc-based threonine-linked polysaccharide with β1,3GlcA side chains [33]. Monoglucuronylated O-glycans with reducing terminal mannose have been described from the fungus Trichoderma reesii [34].
Chondroitin and heparan chains of proteoglycans with varying degrees of sulphation can also be considered as O-glycans, as these primarily consist of HexA1HexNAc1 glycosaminoglycan repeats attached via a GlcAβ1,3Galβ1,3Galβ1,4Xyl tetrasaccharide linker (see Fig. 1) in probably all animals, including cnidarian, annelid, nematode and insect species [35,36,37,38]. The structurally-related dermatan sulphate containing iduronic acid is also found in echinoderms [39], while hyaluronic acid occurring as (GlcAβ3GlcNAcβ4)n repeats is apparently absent from invertebrates [40]. In terms of genetic model organisms, glycosaminoglycans from Caenorhabditis and Drosophila have a number of roles in development as shown by studies on various mutants lacking key enzymes in chondroitin and heparan biosynthesis [41, 42]. Probably the best characterised invertebrate β1,3-glucuronyltransferases are three enzymes from Drosophila with potential to act in N-, O-, and lipid glycan or glycosaminoglycan biosynthesis [43].
Sialic acids
In most invertebrates, it can be safely concluded that sialic acids are absent, as neither the relevant monosaccharides have been detected by modern methods nor the genetic capacity to make or transfer them is present. Insects constitute an exception as these do have sialyltransferase genes; however, only in Drosophila is there proper evidence as to it being attached to glycans [44]. On the other hand, as discussed below, primitive deuterostomes such as echinoderms have multiple sialyltransferase genes and a number of studies have proven the occurrence of sialylated glycans. However, the presence of plant genes with homology to sialyltransferases [45] also throws up questions as to the real enzymatic and biological role of such proteins in organisms for which the occurrence of sialic acid on glycans is not proven; indeed, more recently it has been proposed that these plant sialyltransferase-like enzymes may transfer either 2-keto-3-deoxy-D-lyxo-heptulosaric acid (DHA) or 2-keto-3-deoxy-D-manno-octulosonic acid (KDO) to rhamnogalacturonan-II [46]. In the case of trypanosomatids, trans-sialylation results in parasites coating their mucins with host-derived sialic acid [47]. Various reports claiming that sialic acid is present in other invertebrate organisms, however, must be taken with caution, as mere HPLC peaks or lectin blotting data are not conclusive proofs and can be due to, e.g., lectin cross-reactivity or the presence of contaminants; only mass spectrometric proof of the structure, in the absence of deuterostome components in media or food, is an acceptable demonstration of sialylation.
Sialylation in insects has long been controversial. One early report suggested the presence of polysialic acid in Drosophila [48], but the only sialylated glycans found by mass spectrometry contain a single residue of N-acetylneuraminic acid attached to galactose on N-glycans [44, 49] (see example in Fig. 1). The single Drosophila sialyltransferase has been functionally and biochemically characterised [50, 51]. In other insect species, sialyltransferase genes have been identified and parts of the CMP-sialic acid biosynthesis pathway revealed [52, 53]; however, only upon glycoengineering can it be safely concluded that Lepidoptera will express sialic acid on recombinant glycoproteins [54].
In the deuterostome lineage, even in organisms evolutionarily more primitive than the vertebrates, there is quite definitely an expansion of the sialyltransferase gene family. For instance, multiple sialyltransferases are encoded by the genome of a cephalochordate (Branchiostoma belcheri); both N-acetylneuraminic acid (Neu5Ac) or N-glycolylneuraminic acid (Neu5Gc) were found to be expressed in a tissue-specific manner in this organism, while methylated 3-deoxy-D-glycero-D-galacto-2-nonulosonic acid (KDN; a non-acylated sialic acid) was found on ovary O-glycans [55]. In echinoderms, a number of α2,3-, α2,6- and α2,8-sialyltransferase homologues, yet to be enzymatically characterised, occur in the sea urchin Strongylocentrotus purpuratus [56,57,58]; NeuAc and NeuGc are found in a number of echinoderm species, also in O-acetylated and O-sulphated forms. In starfish and sea cucumbers, a large percentage of these sialic acids are additionally methylated at O-8, whereas in other echinoderms this position can be sulphated [59, 60]. Relevant metabolic enzymes for the generation of methylated NeuGc are the CMP-NeuAc hydroxylase, as also found in non-human mammals, and an 8-O-methyltransferase [61, 62].
Specific examples of defined sialylated structures in echinoderms include a mucin-type O-glycan from the sperm flagella of the sea urchin (Hemicentrotus pulcherrimus) which contains terminal sulphated O-8 α2,9-linked polyNeu5Ac linked to the protein backbone via GalNAc [63, 64], a motif also present in two other sea urchins (Strongylocentrotus purpuratus and Str. franciscanus). On other sea urchin sperm and egg O-linked glycoproteins, α2,8-sialic acid and α2,5-sialic acid were found [65, 66]. Gangliosides (i.e., lactosyl- or glucosylceramides carrying one or more NeuAc or NeuGc residues, reportedly also in fucosylated, methylated or sulphated forms; see example in Fig. 1) represent another class of sialylated glycoconjugates studied in species of various echinoderm classes [67,68,69,70,71], such as sea cucumbers (Stichopus chloronotus, Sti. japonicus and Holothuria leucospilota), starfish (Linckia laevigata, Asterias amurensis, Luidia maculata, Asterina pectinifera and Acanthaster planci), sea urchins (Str. intermedius and H. pulcherrimus), a feather star (Comanthus japonica) and a brittle star (Ophiocoma scolopendrina).
Pyruvylation
The presence of pyruvate on N-glycans is known from yeast species, specifically 4,6-ketal-linked to outer chain galactose residues [72], whereby it should also be noted that some bacterial and seaweed polysaccharides are also pyruvylated. The pvg1 gene, a pyruvyltransferase, is one of five necessary for pyruvylation in the fission yeast, whereby the pvg3 gene encodes the galactosyltransferase required for transfer of the underlying β1,3-linked galactose residue [73, 74]. Pyruvate was also found on glycolipids of the sea hare, which is a mollusc [75] and on the glyconectin polysaccharides isolated from some Porifera sponges [76].
Sulphate
Probably sulphation is most familiar from glycosaminoglycans (e.g., heparan, chondroitin and keratan sulphates), but some of these are either absent or undersulphated in many invertebrates [77]; on the other hand, some marine organisms have highly sulphated glycosaminoglycans, sometimes also with fucosyl modifications [78]. Indeed, an increased degree of sulphation on various glycoconjugates seems to occur in the context of marine or saline environments [79, 80]. Especially glycosaminoglycans from sharks, hagfish, king crabs, squid, molluscs, sea cucumbers and sea squirts are characterized by oversulphated complex structures which carry more sulphated residues per disaccharide unit than the terrestrial vertebrates on their chondroitin and dermatan sulphates [77], which contain only one sulphate group per disaccharide unit (one, two or three sulphates being possible, of course, on the disaccharide units of vertebrate heparan sulphate). Sulphated fucans and galactans from echinoderms, distinct from glycosaminoglycans, may have roles in fertilisation [81, 82]. Also, although sulphated polysaccharides have not been described in vascular plants (angiosperms), sulphated galactans and fuc(oid)ans have been isolated from marine sea grass species and algae [79, 80], while sulphoquinovosyldiacylglycerol is an important anionic glycolipid component of plant chloroplasts [83]. Sulphated O-glycans with mannose at the reducing terminus have been described from a coral [84].
In terms of N-glycans, sulphation of α-linked mannose and/or core α1,6-fucose is known from arthropods (specifically a lobster as well as insects; see examples in Fig. 1) and from Dictyostelium [21, 85,86,87,88], while sulphation of galactose is a feature of the Eastern oyster [89]. Based on studies with radioactive SO4 labelling and PNGase F treatment, the occurrence of sulphated N-linked oligosaccharide chains on glycoproteins involved in sea urchin embryonic skeleton formation was postulated [90]; these glycan chains, recognised by the 1223 antibody, apparently bind calcium [91], but their exact structures were not defined. Sulphated ‘mucin-type’ O-glycans are known from a marine snail as well as from dipteran species, whereby the major structures are based on the core 1 Galβ1,3GalNAc motif [20, 21]. More unusual are the sulphated O-glycans with Hex-HexNAc-HexA repeats found in mosquito larvae and a recombinant protein expressed in HighFive cells [21, 30]. As mentioned above, 8-O-sulphated sialic acid has been found on sea urchin O-glycans and glycolipids [63, 70]. Generally, the sulphate on glycans originates from 3′-phosphoadenosine-5′-phosphosulfate as the activated donor; however, other than sulphotransferases involved in glycosaminoglycan synthesis [92,93,94,95,96], probably no other invertebrate glycan-modifying sulphotransferases have been either biochemically or biologically characterised.
Phosphate and methylphosphate
In mammals, mannose-6-phosphate is well known for its role in trafficking of lysosomal enzymes; defects in the two-step phosphorylation process (first transfer of GlcNAc-1-phosphate in 6-linkage to mannose, then removal of the GlcNAc to reveal mannose-6-phosphate) result in mislocalisation of lysosomal hydrolases and so to forms of lysosomal storage diseases [97]. Three decades ago, GlcNAc-1-phosphotransferase activities were found in amoebae [98]; more recently the relevant gpt1 gene encoding this enzyme from Dictyostelium has been identified [99]. The final glycan structures in Dictyostelium discoideumwild-type and mannosyltransferase mutant strains contained rather methylated phosphate attached to mannose [88, 100] (see Fig. 2). In a glucosidase II amoebal mutant, however, there was also a significant amount of unmodified and GlcNAc-modified phosphate [101, 102], which is indicative that the biosynthetic route to the methylphosphorylated structures is affected by other aspects of glycan processing. A phosphorylated Man4GlcNAc2 was also found in one strain of Trichomonas vaginalis, a human parasite [103]; it is unclear whether this is due to loss of an ethanolamine from a phosphoethanolamine-modified structure otherwise found in this species. Another form of mannosyl phosphorylation is represented by the mannose-phosphodiesters found in hypermannosylated structures from Saccharomyces cerevisiae and originates by transfer from GDP-Man rather than by a kinase reaction [104, 105]; other phosphodiester-type glycans include the phosphoglycan on the gp72 of Trypanosoma cruzi, the polyhexose chains on the lipoproteophosphoglycan of Entamoeba histolytica and the GlcNAc-P-Ser motif on proteins from Dictyostelium discoideum [106,107,108] (for the latter two examples, see Fig. 2). In terms of analyses, phosphorylated glycans are well observed in positive and negative modes of mass spectrometry and the phosphates can be removed by hydrofluoric acid or (if not methylated) by phosphatases [16].
Example zwitterionic and phosphodiester structures. The depicted structures are (i) a methylphosphorylated N-glycan structure from Dictyostelium discoideum, (ii) an N-glycan from Lymantria dispar modified with glucuronic acid and phosphorylcholine, (iii) a schematic honeybee N-glycan, (iv) a schematic glycan from either nematode or cestode species, (v) a phosphoethanolamine-modified N-glycan from Penicillium spp. optionally with an ‘outer chain’ mannose and a bisecting galactofuranose, (vi) two N-glycans with methylaminoethylphosphonate or phosphorylcholine from Volvarina rubella, (vii) glycolipids from insects (example from Drosophila), nematodes and annelids (with phosphorylcholine) and molluscs (with aminoethylphosphonate; also observed with pyruvate and phosphoethanolamine modifications), (viii) a glycosylphosphatidylinositol (GPI) anchor from the Trypanosoma cruzi NETNES protein, (ix) a mucin-type O-glycan from wasp, a mucin-type and a Notch-type from Volvarina rubella, a glycosaminoglycan-like glycan from Oesophagostomum dentatum and (x) phospho-linked sugars from either Dictyostelium discoideum or Entamoeba histolytica. AEP, aminoethylphosphonate; MeAEP, methylaminoethylphosphonate; P, phosphate; PC, phosphorylcholine; PE, phosphoethanolamine; PMe, methylphosphate; Pyr, pyruvate
Phosphorylcholine and phosphoethanolamine
Other than phosphate, zwitterionic phosphodiesters modify glycoconjugates from a range of prokaryotic and eukaryotic organisms. Phosphoethanolamine (PE; also known as aminoethylphosphate or ethanolamine phosphate; Δm/z 123 Da) and its tri-N-methylated form, phosphorylcholine (PC; Δm/z 165 Da), have been described on N-glycans, O-glycans, glycolipids and glycosaminoglycans from protists, fungi, insects, annelids and nematodes as well as on lipopolysaccharides from various bacteria. Phosphoethanolamine is also a component of eukaryotic glycosylphosphatidylinositol (GPI) anchors [109].
In nematodes and in one cestode it appears that GlcNAc or GalNAc residues of N-glycans and glycolipids can be modified with phosphorylcholine (see examples in Fig. 2), as shown by studies on C. elegans, Ascaris suum, Trichuris suis, Haemonchus contortus, Oesophagostomum dentatum, Pristionchus pacificus, Trichinella spiralis, various filarial worms (including Dirofilaria immitis) and Echinococcus granulosus [25, 110,111,112,113,114,115,116,117,118,119]; the underlying structures of the phosphorylcholine-modified N-glycans from nematodes vary, but multiple residues (in the context of a single GlcNAc, chito-oligomer or LacdiNAc-type motifs) on two, three or four antennae are possible. Phosphorylcholine is also found on annelid and nematode glycolipids [120,121,122,123], on glycosaminoglycan-like structures from at least one nematode [124] and on O-glycans with the composition HexNAc2-3HexA1PC1 released from a recombinant protein expressed in lepidopteran (Sf9) cells [30]. In fungi, phosphorylcholine is found to substitute either galactofuranose on glycosylinositol-phosphoceramide glycolipids [125] or mannose in a peptidophosphogalactomannan [126].
Phosphorylcholine was found on core ‘GalFuc’ moieties of N-glycans of the gastropod Volvarina rubella [20] as well as on the antennae of a not insignificant modification of N-glycans from Lepidoptera, including cell lines used in biotechnology such as Trichoplusia ni HighFive [24]. In other insects, it is rather phosphoethanolamine that is found on N-glycans of the honeybee [23] and the O-glycans of wasps [127]; on the other hand, N-glycans from Diptera were not yet found to contain zwitterionic modifications, even though phosphoethanolamine linked to GlcNAc is a component of glycolipids from dipteran insects such as Drosophila and Calliphora [128, 129] (see example in Fig. 2) or on O-glycans from mosquito larvae with the composition HexNAc1-2Hex1HexA2PE1 [21]. Phosphoethanolamine is also present on mannose residues of N-glycans from Penicillium species and Trichomonas vaginalis [103, 130] and has been reported as a modification of glycolipids from the bivalve, Corbicula sandai [131].
At the analytical level, phosphoethanolamine yields signals in both positive and negative modes of MALDI-TOF MS, while the phosphorylcholine is only (and excellently) detectable in positive mode [16]; however, both are sensitive to hydrofluoric acid hydrolysis. The enzymatic basis for addition of these moieties is far from resolved, but conceivably CDP-choline/ CDP-ethanolamine or phosphatidylcholine/ phosphatidylethanolamine could be the donor substrates for PC/PE-transferases [132,133,134]. Both phosphorylcholine and phosphoethanolamine are ligands for pentraxins (e.g., human C-reactive protein and serum amyloid P), which are components of the innate immune system [135]; at least phosphorylcholine modifications on glycolipids and glycoproteins are associated with immunomodulatory activity, as especially shown for the Acanthocheilonema viteaeES-62 protein, which is an excretory-secretory protein of a rodent nematode parasite, whereby the Toll-like receptor 4 (TLR4) may be involved in the relevant signalling pathway [136].
Aminoethylphosphonate
Phosphonates differ from phosphates in that there is a carbon-phosphorus bond without any intermediate oxygen atom. In eukaryotes, the first demonstrations of the 2-aminoethylphosphonate and N-methyl-2-aminoethylphosphonate modifications of glycans (Δm/z 107 and 121 Da) came from studies on glycolipids of gastropods, such as the sea hare Aplysia kurodai [137,138,139,140,141,142]; as shown in Fig. 2, one such glycolipid is reported to also contain pyruvate and phosphoethanolamine residues [75]. In other studies, a GPI anchor from Trypanosoma cruzi (the causative agent of Chagas’ disease), N-glycans from locust and O-glycans from jellyfish were found to be modified with aminoethylphosphonate [143,144,145]. Most recently, methylaminoethylphosphonate and aminoethylphosphonate were detected on N-glycans as well as O-fucose-based structures from a marine snail [20], while there is also evidence that oligomannosidic glycans from Euglena gracilis, a free-living protist, carry aminoethylphosphonate [145].
Predominantly, it seems that either mannose or GlcNAc residues can be modified by these phosphonates; like phosphorylcholine and phosphoethanolamine, these can be cleaved by hydrofluoric acid. Aminoethylphosphonates were detected in both positive and negative ion modes of FAB- and MALDI-TOF-MS. Neither the recognition of aminoethylphosphonate by pentraxins nor the routes for its transfer to glycan substrates is known.
Mass alone is not enough
Perhaps part of the reason for the historically relatively low numbers of proven anionic and zwitterionic glycans in non-vertebrates is that they were not expected. Also, in screens based on positive mode mass spectrometry alone without fragmentation, many such structures would not be noticed as they are isobaric (i.e., same mass) as compared to more well-known structures; most commonly, a typical mass spectrometer without a very high resolution will not distinguish a phosphate from a sulphate. Combined with enzymatic or chemical treatments of glycans, careful assessment of MS/MS fragments is indeed required, as perhaps only a couple of fragments are showing the structural difference. For instance, MS/MS is necessary to show that a neutral complex N-glycan is not actually carrying phosphoethanolamine (see examples for m/z 2004, 2328 and 2870; Fig. 3 a-f) or a glycan with the same mass as a oligomannosidic glycan is actually one modified with methylaminoethylphosphonate on antennal GlcNAc (see the examples for m/z 1151 and 1637; Fig. 3 j-o); other examples offered by the marine snail Volvarina rubella include (i) a glycan with four methylaminoethylphosphonate residues isobaric (m/z 1879; Fig. 3 p) with glucuronylated structures from insects (Fig. 3 q and r) or (ii) a glycan with a bisubstituted fucose residue with the same mass (m/z 1852; Fig. 3 s) as structures from insects or nematodes carrying phosphorylcholine (Fig. 3t and u). As we have previously reviewed in this journal [16], not only separation of classes and isomers of glycans is required, the different ionisation and fragmentation in positive and negative modes of mass spectrometry is useful in showing whether a glycan is indeed that ‘predicted’ from the mass, while hydrofluoric acid hydrolysis is valuable as it will remove phosphate esters (Fig. 3 g-i), fucose (especially in α1,3-linkage) and galactofuranose, but not sulphate or other hexoses.
Mass spectrometry of isobaric/isomeric structures. a-f Example positive mode MS/MS of N-glycans from Apis mellifera royal jelly or larvae which isobarically differ depending on the presence of phosphoethanolamine, LacdiNAc or glucuronylated motifs. g-i Example positive (+) or negative (−) mode MS of isobaric glycans from an echinoderm or Volvarina with masses of 1271 Da carrying either phosphate, sulphate (note in-source loss in positive mode) or methylaminoethylphosphonate moieties and are distinguishable due to their ionisation in positive mode or sensitivity to hydrofluoric acid (HF). j-l Positive mode MS/MS of isobaric/isomeric variations of 1150 Da (m/z 1151) from Volvarina rubella either methylaminoethylphosphonate-modified, core β-mannosylated or ‘normal’ paucimannosidic. m-u Positive mode MS/MS of variations of glycans of m/z 1637, 1879 or 1852 which are either methylaminoethylphosphonate-modified (Volvarina), glucuronylated (Apis), phosphorylcholine-modified (Trichoplusia or Dirofilaria) or standard oligomannosidic structures. Annotated are the key fragments (symbolic nomenclature), losses (with arrows) or cleavages (red bars and m/z values); all glycans were reductively aminated with 2-aminopyridine (PA) which yields typical reducing terminal Y fragments of m/z 300, 446 or 462 (GlcNAc1Fuc0–1Man0–1-PA). Abbreviated compositions of the form HxNyF0–1 U0–1 correspond to HexxHexNAcyFuc0–1HexA0–1; MEAP (or *), methylaminoethylphosphonate; PC, phosphorylcholine; PE, phosphoethanolamine; S, sulphate
Conclusion
In this brief overview of anionic and zwitterionic glycans from various lower eukaryotes, it is obvious that the glycosylation in protists and invertebrates is highly variable. Of course, our knowledge is only based on those organisms or phyla which have been investigated; very many phyla (and naturally huge numbers of species) have never been studied (or adequately so) as to their glycomes. Mere mass spectrometric profiling, without doing deep analyses based on fractionated glycomes, will also not show the true extent of the glycostructural possibilities. As many anionic or zwitterionic glycans with potential biological activity occur in low amounts, they are often overseen if, e.g., the whole N-glycome is ‘shot’ from a single spot on a MALDI plate; certainly the isomeric structures will not be easily distinguished. However, as technologies and methodologies evolve, there is potential for even more variants and/or commonalities between species.
References
Varki, A.: Nothing in glycobiology makes sense, except in the light of evolution. Cell. 126, 841–845 (2006). https://doi.org/10.1016/j.cell.2006.08.022
Schauer, R., Kamerling, J.P.: Exploration of the sialic acid world. Adv. Carbohydr. Chem. Biochem. 75, 1–213 (2018). https://doi.org/10.1016/bs.accb.2018.09.001
Bülow, H.E., Hobert, O.: The molecular diversity of glycosaminoglycans shapes animal development. Annu. Rev. Cell Dev. Biol. 22, 375–407 (2006). https://doi.org/10.1146/annurev.cellbio.22.010605.093433
Morita, I., Kizuka, Y., Kakuda, S., Oka, S.: Expression and function of the HNK-1 carbohydrate. J. Biochem. 143, 719–724 (2008). https://doi.org/10.1093/jb/mvm221
Honke, K., Taniguchi, N.: Sulfotransferases and sulfated oligosaccharides. Med. Res. Rev. 22, 637–654 (2002). https://doi.org/10.1002/med.10020
Pomin, V.H.: Phylogeny, structure, function, biosynthesis and evolution of sulfated galactose-containing glycans. Int. J. Biol. Macromol. 84, 372–379 (2016). https://doi.org/10.1016/j.ijbiomac.2015.12.035
Pohl, S., Marschner, K., Storch, S., Braulke, T.: Glycosylation- and phosphorylation-dependent intracellular transport of lysosomal hydrolases. Biol. Chem. 390(521–527), (2009). https://doi.org/10.1515/BC.2009.076
Kinoshita, T., Fujita, M.: Biosynthesis of GPI-anchored proteins: special emphasis on GPI lipid remodeling. J. Lipid Res. 57, 6–24 (2016). https://doi.org/10.1194/jlr.R063313
Praissman, J.L., Willer, T., Sheikh, M.O., Toi, A., Chitayat, D., Lin, Y.Y., Lee, H., Stalnaker, S.H., Wang, S., Prabhakar, P.K., Nelson, S.F., Stemple, D.L., Moore, S.A., Moremen, K.W., Campbell, K.P., Wells, L.: The functional O-mannose glycan on α-dystroglycan contains a phospho-ribitol primed for matriglycan addition. Elife. 5, e14473 (2016). https://doi.org/10.7554/eLife.14473
Scheller, H.V., Ulvskov, P.: Hemicelluloses. Annu. Rev. Plant Biol. 61, 263–289 (2010). https://doi.org/10.1146/annurev-arplant-042809-112315
Caffall, K.H., Mohnen, D.: The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 344, 1879–1900 (2009). https://doi.org/10.1016/j.carres.2009.05.021
Li, W., De Schutter, K., Van Damme, E.J.M., Smagghe, G.: Synthesis and biological roles of O-glycans in insects. Glycoconj. J. (2019). https://doi.org/10.1007/s10719-019-09867-1
Paschinger, K., Yan, S., Wilson, I.B.H.: N-glycomic complexity in anatomical simplicity: Caenorhabditis elegans as a non-model nematode? Front. Mol. Biosci. 6(9), (2019). https://doi.org/10.3389/fmolb.2019.00009
Zhu, F., Li, D., Chen, K.: Structures and functions of invertebrate glycosylation. Open Biol. 9, 180232 (2019). https://doi.org/10.1098/rsob.180232
Hwang, H.-Y., Horvitz, H.R.: The Caenorhabditis elegans vulval morphogenesis gene sqv-4 encodes a UDP-glucose dehydrogenase that is temporally and spatially regulated. Proc. Natl. Acad. Sci. U. S. A. 99, 14224–14229 (2002)
Paschinger, K., Wilson, I.B.H.: Analysis of zwitterionic and anionic N-linked glycans from invertebrates and protists by mass spectrometry. Glycoconj. J. 33, 273–283 (2016). https://doi.org/10.1007/s10719-016-9650-x
Caffall, K.H., Pattathil, S., Phillips, S.E., Hahn, M.G., Mohnen, D.: Arabidopsis thaliana T-DNA mutants implicate GAUT genes in the biosynthesis of pectin and xylan in cell walls and seed testa. Mol. Plant. 2, 1000–1014 (2009). https://doi.org/10.1093/mp/ssp062
Dejima, K., Takemura, M., Nakato, E., Peterson, J., Hayashi, Y., Kinoshita-Toyoda, A., Toyoda, H., Nakato, H.: Analysis of Drosophila glucuronyl C5-epimerase: implications for developmental roles of heparan sulfate sulfation compensation and 2-O-sulfated glucuronic acid. J. Biol. Chem. 288, 34384–34393 (2013). https://doi.org/10.1074/jbc.M113.499269
Zhou, H., Hanneman, A.J., Chasteen, N.D., Reinhold, V.N.: Anomalous N-glycan structures with an internal fucose branched to GlcA and GlcN residues isolated from a mollusk shell-forming fluid. J. Proteome Res. 12, 4547–4555 (2013). https://doi.org/10.1021/pr4006734
Eckmair, B., Jin, C., Abed-Navandi, D., Paschinger, K.: Multi-step fractionation and mass spectrometry reveals zwitterionic and anionic modifications of the N- and O-glycans of a marine snail. Mol. Cell. Proteomics. 15, 573–597 (2016). https://doi.org/10.1074/mcp.M115.051573
Kurz, S., Aoki, K., Jin, C., Karlsson, N.G., Tiemeyer, M., Wilson, I.B.H., Paschinger, K.: Targetted release and fractionation reveal glucuronylated and sulphated N- and O-glycans in larvae of dipteran insects. J. Proteome. 126, 172–188 (2015). https://doi.org/10.1016/j.jprot.2015.05.030
Aoki, K., Tiemeyer, M.: The glycomics of glycan glucuronylation in Drosophila melanogaster. Methods Enzymol. 480, 297–321 (2010). https://doi.org/10.1016/S0076-6879(10)80014-X
Hykollari, A., Malzl, D., Eckmair, B., Vanbeselaere, J., Scheidl, P., Jin, C., Karlsson, N.G., Wilson, I.B.H., Paschinger, K.: Isomeric separation and recognition of anionic and zwitterionic N-glycans from royal jelly glycoproteins. Mol. Cell. Proteomics. 17, 2177–2196 (2018). https://doi.org/10.1074/mcp.RA117.000462
Stanton, R., Hykollari, A., Eckmair, B., Malzl, D., Dragosits, M., Palmberger, D., Wang, P., Wilson, I.B.H., Paschinger, K.: The underestimated N-glycomes of lepidopteran species. Biochim. Biophys. Acta. 1861, 699–714 (2017). https://doi.org/10.1016/j.bbagen.2017.01.009
Martini, F., Eckmair, B., Neupert, C., Štefanić, S., Jin, C., Garg, M., Jiménez-Castells, C., Hykollari, A., Yan, S., Venco, L., Varón Silva, D., Wilson, I.B.H., Paschinger, K.: Highly modified and immunoactive N-glycans of the canine heartworm. Nat. Commun. 10(75), 75 (2019). https://doi.org/10.1038/s41467-018-07948-7
Wiegandt, H.: Insect glycolipids. Biochim. Biophys. Acta. 1123, 117–126 (1992)
Fontaine, T., Lamarre, C., Simenel, C., Lambou, K., Coddeville, B., Delepierre, M., Latge, J.P.: Characterization of glucuronic acid containing glycolipid in Aspergillus fumigatus mycelium. Carbohydr. Res. 344, 1960–1967 (2009). https://doi.org/10.1016/j.carres.2009.07.012
Heiss, C., Skowyra, M.L., Liu, H., Klutts, J.S., Wang, Z., Williams, M., Srikanta, D., Beverley, S.M., Azadi, P., Doering, T.L.: Unusual galactofuranose modification of a capsule polysaccharide in the pathogenic yeast Cryptococcus neoformans. J. Biol. Chem. 288, 10994–11003 (2013). https://doi.org/10.1074/jbc.M112.441998
Breloy, I., Schwientek, T., Lehr, S., Hanisch, F.G.: Glucuronic acid can extend O-linked core 1 glycans, but it contributes only weakly to the negative surface charge of Drosophila melanogasterSchneider-2 cells. FEBS Lett. 582, 1593–1598 (2008). https://doi.org/10.1016/j.febslet.2008.04.003
Gaunitz, S., Jin, C., Nilsson, A., Liu, J., Karlsson, N.G., Holgersson, J.: Mucin-type proteins produced in the Trichoplusia ni and Spodoptera frugiperda insect cell lines carry novel O-glycans with phosphocholine and sulfate substitutions. Glycobiology. 23, 778–796 (2013). https://doi.org/10.1093/glycob/cwt015
Itoh, K., Akimoto, Y., Kondo, S., Ichimiya, T., Aoki, K., Tiemeyer, M., Nishihara, S.: Glucuronylated core 1 glycans are required for precise localization of neuromuscular junctions and normal formation of basement membranes on Drosophila muscles. Dev. Biol. 436, 108–124 (2018). https://doi.org/10.1016/j.ydbio.2018.02.017
Palaima, E., Leymarie, N., Stroud, D., Mizanur, R.M., Hodgkin, J., Gravato-Nobre, M.J., Costello, C.E., Cipollo, J.F.: The Caenorhabditis elegans bus-2 mutant reveals a new class of O-glycans affecting bacterial resistance. J. Biol. Chem. 285, 17662–17672 (2010). https://doi.org/10.1074/jbc.M109.065433
Bergwerff, A.A., Van Dam, G.J., Rotmans, J.P., Deelder, A.M., Kamerling, J.P., Vliegenthart, J.F.G.: The immunologically reactive part of immunopurified circulating anodic antigen from Schistosoma mansoni is a threonine- linked polysaccharide consisting of →6)-(β-D-GlcpA-(1→3))-β-D-GalpNAc-(1→ repeating units. J. Biol. Chem. 269, 31510–31517 (1994)
Ashwood, C., Abrahams, J.L., Nevalainen, H., Packer, N.H.: Enhancing structural characterisation of glucuronidated O-linked glycans using negative mode ion trap higher energy collision-induced dissociation mass spectrometry. Rapid Commun. Mass Spectrom. 31, 851–858 (2017). https://doi.org/10.1002/rcm.7851
Yamada, S., Okada, Y., Ueno, M., Iwata, S., Deepa, S.S., Nishimura, S., Fujita, M., Van Die, I., Hirabayashi, Y., Sugahara, K.: Determination of the glycosaminoglycan-protein linkage region oligosaccharide structures of proteoglycans from Drosophila melanogaster and Caenorhabditis elegans. J. Biol. Chem. 277, 31877–31886 (2002)
Lawrence, R., Olson, S.K., Steele, R.E., Wang, L., Warrior, R., Cummings, R.D., Esko, J.D.: Evolutionary differences in glycosaminoglycan fine structure detected by quantitative glycan reductive isotope labeling. J. Biol. Chem. 283, 33674–33684 (2008). https://doi.org/10.1074/jbc.M804288200
Im, A.R., Park, Y., Sim, J.S., Zhang, Z., Liu, Z., Linhardt, R.J., Kim, Y.S.: Glycosaminoglycans from earthworms (Eisenia andrei). Glycoconj. J. 27, 249–257 (2010). https://doi.org/10.1007/s10719-009-9273-6
Noborn, F., Gomez Toledo, A., Nasir, W., Nilsson, J., Dierker, T., Kjellen, L., Larson, G.: Expanding the chondroitin glycoproteome of Caenorhabditis elegans. J. Biol. Chem. 293, 379–389 (2018). https://doi.org/10.1074/jbc.M117.807800
Vilela-Silva, A.C., Werneck, C.C., Valente, A.P., Vacquier, V.D., Mourão, P.A.: Embryos of the sea urchin Strongylocentrotus purpuratus synthesize a dermatan sulfate enriched in 4-O- and 6-O-disulfated galactosamine units. Glycobiology. 11, 433–440 (2001)
Csoka, A.B., Stern, R.: Hypotheses on the evolution of hyaluronan: a highly ironic acid. Glycobiology. 23, 398–411 (2013). https://doi.org/10.1093/glycob/cws218
Hwang, H.-Y., Olson, S.K., Esko, J.D., Horvitz, H.R.: Caenorhabditis elegans early embryogenesis and vulval morphogenesis require chondroitin biosynthesis. Nature. 423, 439–443 (2003)
Izumikawa, T., Egusa, N., Taniguchi, F., Sugahara, K., Kitagawa, H.: Heparan sulfate polymerization in Drosophila. J. Biol. Chem. 281, 1929–1934 (2006). https://doi.org/10.1074/jbc.M509138200
Kim, B.-T., Tsuchida, K., Lincecum, J., Kitagawa, H., Bernfield, M., Sugahara, K.: Identification and characterization of three Drosophila melanogaster glucuronyltransferases responsible for the synthesis of the conserved glycosaminoglycan-protein linkage region of proteoglycans. Two novel homologs exhibit broad specificity toward oligosaccharides from proteoglycans, glycoproteins, and glycosphingolipids. J. Biol. Chem. 278, 9116–9124 (2003). https://doi.org/10.1074/jbc.M209344200
Aoki, K., Perlman, M., Lim, J.M., Cantu, R., Wells, L., Tiemeyer, M.: Dynamic developmental elaboration of N-linked glycan complexity in the Drosophila melanogaster embryo. J. Biol. Chem. 282, 9127–9142 (2007)
Takashima, S., Abe, T., Yoshida, S., Kawahigashi, H., Saito, T., Tsuji, S., Tsujimoto, M.: Analysis of Sialyltransferase-like proteins from Oryza sativa. J Biochem (Tokyo). 139, 279–287 (2006)
Dumont, M., Lehner, A., Bouton, S., Kiefer-Meyer, M.C., Voxeur, A., Pelloux, J., Lerouge, P., Mollet, J.C.: The cell wall pectic polymer rhamnogalacturonan-II is required for proper pollen tube elongation: implications of a putative sialyltransferase-like protein. Ann. Bot. 114, 1177–1188 (2014). https://doi.org/10.1093/aob/mcu093
Eugenia Giorgi, M., de Lederkremer, R.M.: Trans-sialidase and mucins of Trypanosoma cruzi: an important interplay for the parasite. Carbohydr. Res. 346, 1389–1393 (2011). https://doi.org/10.1016/j.carres.2011.04.006
Roth, J., Kempf, A., Reuter, G., Schauer, R., Gehring, W.J.: Occurrence of sialic acids in Drosophila melanogaster. Science. 256, 673–675 (1992)
Frappaolo, A., Sechi, S., Kumagai, T., Robinson, S., Fraschini, R., Karimpour-Ghahnavieh, A., Belloni, G., Piergentili, R., Tiemeyer, K.H., Tiemeyer, M., Giansanti, M.G.: COG7 deficiency in Drosophila generates multifaceted developmental, behavioral and protein glycosylation phenotypes. J. Cell Sci. 130, 3637–3649 (2017). https://doi.org/10.1242/jcs.209049
Koles, K., Irvine, K.D., Panin, V.M.: Functional characterization of Drosophila sialyltransferase. J. Biol. Chem. 279, 4346–4357 (2004)
Repnikova, E., Koles, K., Nakamura, M., Pitts, J., Li, H., Ambavane, A., Zoran, M.J., Panin, V.M.: Sialyltransferase regulates nervous system function in Drosophila. J. Neurosci. 30, 6466–6476 (2010). https://doi.org/10.1523/JNEUROSCI.5253-09.2010
Kajiura, H., Hamaguchi, Y., Mizushima, H., Misaki, R., Fujiyama, K.: Sialylation potentials of the silkworm, Bombyx mori; B. mori possesses an active α2,6-sialyltransferase. Glycobiology. 25, 1441–1453 (2015). https://doi.org/10.1093/glycob/cwv060
Di, W., Fujita, A., Hamaguchi, K., Delannoy, P., Sato, C., Kitajima, K.: Diverse subcellular localizations of the insect CMP-sialic acid synthetases. Glycobiology. 27, 329–341 (2017). https://doi.org/10.1093/glycob/cww128
Aumiller, J.J., Hollister, J., Jarvis, D.L.: A transgenic insect cell line engineered to produce CMP-sialic acid and sialylated glycoproteins. Glycobiology. 13, 497–507 (2003). https://doi.org/10.1093/glycob/cwg051
Guérardel, Y., Chang, L.Y., Fujita, A., Coddeville, B., Maes, E., Sato, C., Harduin-Lepers, A., Kubokawa, K., Kitajima, K.: Sialome analysis of the cephalochordate Branchiostoma belcheri, a key organism for vertebrate evolution. Glycobiology. 22, 479–491 (2012)
Harduin-Lepers, A., Mollicone, R., Delannoy, P., Oriol, R.: The animal sialyltransferases and sialyltransferase-related genes: a phylogenetic approach. Glycobiology. 15, 805–817 (2005). https://doi.org/10.1093/glycob/cwi063
Petit, D., Teppa, E., Mir, A.M., Vicogne, D., Thisse, C., Thisse, B., Filloux, C., Harduin-Lepers, A.: Integrative view of α2,3-sialyltransferases (ST3Gal) molecular and functional evolution in deuterostomes: significance of lineage-specific losses. Mol. Biol. Evol. 32, 906–927 (2015). https://doi.org/10.1093/molbev/msu395
Harduin-Lepers, A., Petit, D., Mollicone, R., Delannoy, P., Petit, J.M., Oriol, R.: Evolutionary history of the α2,8-sialyltransferase (ST8Sia) gene family: tandem duplications in early deuterostomes explain most of the diversity found in the vertebrate ST8Sia genes. BMC Evol. Biol. 8, 258 (2008). https://doi.org/10.1186/1471-2148-8-258
Warren, L.: N-Glycolyl-8-O-Methylneuraminic acid, a new form of sialic acid in the starfish Asterias forbesi. Biochim. Biophys. Acta. 83, 129–132 (1964)
Klein, A., Diaz, S., Ferreira, I., Lamblin, G., Roussel, P., Manzi, A.E.: New sialic acids from biological sources identified by a comprehensive and sensitive approach: liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS) of SIA quinoxalinones. Glycobiology. 7, 421–432 (1997)
Schlenzka, W., Shaw, L., Schauer, R.: Catalytic properties of the CMP-N-acetylneuraminic acid hydroxylase from the starfish Asterias rubens: comparison with the mammalian enzyme. Biochim. Biophys. Acta. 1161, 131–138 (1993)
Kelm, A., Shaw, L., Schauer, R., Reuter, G.: The biosynthesis of 8-O-methylated sialic acids in the starfish Asterias rubens- isolation and characterisation of S-adenosyl-L-methionine:sialate-8-O-methyltransferase. Eur. J. Biochem. 251, 874–884 (1998)
Miyata, S., Sato, C., Kumita, H., Toriyama, M., Vacquier, V.D., Kitajima, K.: Flagellasialin: a novel sulfated α2,9-linked polysialic acid glycoprotein of sea urchin sperm flagella. Glycobiology. 16, 1229–1241 (2006)
Miyata, S., Sato, C., Kitamura, S., Toriyama, M., Kitajima, K.: A major flagellum sialoglycoprotein in sea urchin sperm contains a novel polysialic acid, an α2,9-linked poly-N-acetylneuraminic acid chain, capped by an 8-O-sulfated sialic acid residue. Glycobiology. 14, 827–840 (2004). https://doi.org/10.1093/glycob/cwh100
Miyata, S., Yamakawa, N., Toriyama, M., Sato, C., Kitajima, K.: Co-expression of two distinct polysialic acids, α2,8- and α2,9-linked polymers of N-acetylneuraminic acid, in distinct glycoproteins and glycolipids in sea urchin sperm. Glycobiology. 21, 1596–1605 (2011)
Kitazume, S., Kitajima, K., Inoue, S., Haslam, S.M., Morris, H.R., Dell, A., Lennarz, W.J., Inoue, Y.: The occurrence of novel 9-O-sulfatedN-glycolylneuraminic acid-capped α2→5-Oglycolyl-linked oligo/polyNeu5Gc chains in sea urchin egg cell surface glycoprotein. Identification of a new chain termination signal for polysialyltransferase. J. Biol. Chem. 271, 6694–6701 (1996)
Yamada, K., Hamada, A., Kisa, F., Miyamoto, T., Higuchi, R.: Constituents of Holothuroidea, 13. Structure of neuritogenic active ganglioside molecular species from the sea cucumber Stichopus chloronotus. Chem Pharm Bull (Tokyo). 51, 46–52 (2003)
Inagaki, M., Miyamoto, T., Isobe, R., Higuchi, R.: Biologically active glycosides from asteroidea, 43. Isolation and structure of a new neuritogenic-active ganglioside molecular species from the starfish Linckia laevigata. Chem Pharm Bull (Tokyo). 53, 1551–1554 (2005)
Prokazova, N.V., Mikhailov, A.T., Kocharov, S.L., Malchenko, L.A., Zvezdina, N.D., Buznikov, G., Bergelson, L.D.: Unusual gangliosides of eggs and embryos of the sea urchin Strongylocentrotus intermedius. Structure and density-dependence of surface localization. Eur. J. Biochem. 115, 671–677 (1981)
Ijuin, T., Kitajima, K., Song, Y., Kitazume, S., Inoue, S., Haslam, S.M., Morris, H.R., Dell, A., Inoue, Y.: Isolation and identification of novel sulfated and nonsulfated oligosialyl glycosphingolipids from sea urchin sperm. Glycoconj. J. 13, 401–413 (1996)
Higuchi, R., Inagaki, M., Yamada, K., Miyamoto, T.: Biologically active gangliosides from echinoderms. J. Nat. Med. 61, 367–370 (2007). https://doi.org/10.1007/s11418-007-0171-6
Gemmill, T.R., Trimble, R.B.: Schizosaccharomyces pombe produces novel pyruvate-containing N-linked oligosaccharides. J. Biol. Chem. 271, 25945–25949 (1996)
Andreishcheva, E.N., Kunkel, J.P., Gemmill, T.R., Trimble, R.B.: Five genes involved in biosynthesis of the pyruvylated Galβ1,3-epitope in Schizosaccharomyces pombeN-linked glycans. J. Biol. Chem. 279, 35644–35655 (2004). https://doi.org/10.1074/jbc.M403574200
Yoritsune, K., Matsuzawa, T., Ohashi, T., Takegawa, K.: The fission yeast Pvg1p has galactose-specific pyruvyltransferase activity. FEBS Lett. 587, 917–921 (2013). https://doi.org/10.1016/j.febslet.2013.02.016
Araki, S., Abe, S., Yamada, S., Satake, M., Fujiwara, N., Kon, K., Ando, S.: Characterization of two novel pyruvylated glycosphingolipids containing 2′-aminoethylphosphoryl(⟶6)-galactose from the nervous system of Aplysia kurodai. J. Biochem. 112, 461–469 (1992)
Guérardel, Y., Czeszak, X., Sumanovski, L.T., Karamanos, Y., Popescu, O., Strecker, G., Misevic, G.N.: Molecular fingerprinting of carbohydrate structure phenotypes of three porifera proteoglycan-like glyconectins. J. Biol. Chem. 279, 15591–15603 (2004). https://doi.org/10.1074/jbc.M308928200
Yamada, S., Sugahara, K., Ozbek, S.: Evolution of glycosaminoglycans: comparative biochemical study. Commun Integr Biol. 4, 150–158 (2011). https://doi.org/10.4161/cib.4.2.14547
Kariya, Y., Watabe, S., Kyogashima, M., Ishihara, M., Ishii, T.: Structure of fucose branches in the glycosaminoglycan from the body wall of the sea cucumber Stichopus japonicus. Carbohydr. Res. 297, 273–279 (1997)
Aquino, R.S., Grativol, C., Mourão, P.A.: Rising from the sea: correlations between sulfated polysaccharides and salinity in plants. PLoS One. 6, e18862 (2011)
Vasconcelos, A.A., Pomin, V.H.: The sea as a rich source of structurally unique Glycosaminoglycans and mimetics. Microorganisms. 5, (2017). https://doi.org/10.3390/microorganisms5030051
Vilela-Silva, A.C., Hirohashi, N., Mourao, P.A.: The structure of sulfated polysaccharides ensures a carbohydrate-based mechanism for species recognition during sea urchin fertilization. Int J Dev Biol. 52, 551–559 (2008). https://doi.org/10.1387/ijdb.072531av
Pomin, V.H.: Structural and functional insights into sulfated galactans: a systematic review. Glycoconj. J. 27, 1–12 (2010). https://doi.org/10.1007/s10719-009-9251-z
Yu, B., Benning, C.: Anionic lipids are required for chloroplast structure and function in Arabidopsis. Plant J. 36, 762–770 (2003)
Coddeville, B., Maes, E., Ferrier-Pages, C., Guerardel, Y.: Glycan profiling of gel forming mucus layer from the scleractinian symbiotic coral Oculina arbuscula. Biomacromolecules. 12, 2064–2073 (2011). https://doi.org/10.1021/bm101557v
van Kuik, J.A., Breg, J., Kolsteeg, C.E.M., Kamerling, J.P., Vliegenthart, J.F.G.: Primary structure of the acidic carbohydrate chain of Hemocyanin from Panulirus interruptus. FEBS Lett. 221, 150–154 (1987). https://doi.org/10.1016/0014-5793(87)80370-8
Cabrera, G., Salazar, V., Montesino, R., Tambara, Y., Struwe, W.B., Lugo, E.L., Harvey, D.J., Antoine, L., Rincon, M., Domon, B., Mendez Martinez, M.D., Portela, M., Gonzalez-Hernandez, A., Triguero, A., Duran, R., Lundberg, U., Vonasek, E., Gonzalez, L.J.: Structural characterization and biological implications of sulfated N-glycans in a serine protease from the neotropical moth Hylesia metabus (Cramer [1775]) (Lepidoptera: Saturniidae). Glycobiology. 26(230–250), cwv096 (2015). https://doi.org/10.1093/glycob/cwv096
Freeze, H.H.: Mannose 6-sulfate is present in the N-linked oligosaccharides of lysosomal enzymes of Dictyostelium. Arch. Biochem. Biophys. 243, 690–693 (1985)
Hykollari, A., Balog, C.I., Rendic, D., Braulke, T., Wilson, I.B., Paschinger, K.: Mass spectrometric analysis of neutral and anionic N-glycans from a Dictyostelium discoideum model for human congenital disorder of glycosylation CDG IL. J. Proteome Res. 12, 1173–1187 (2013). https://doi.org/10.1021/pr300806b
Kurz, S., Jin, C., Hykollari, A., Gregorich, D., Giomarelli, B., Vasta, G.R., Wilson, I.B.H., Paschinger, K.: Haemocytes and plasma of the eastern oyster (Crassostrea virginica) display a diverse repertoire of sulphated and blood group A-modified N-glycans. J. Biol. Chem. 288, 24410–24428 (2013). https://doi.org/10.1074/jbc.M113.478933
Kabakoff, B., Hwang, S.P., Lennarz, W.J.: Characterization of post-translational modifications common to three primary mesenchyme cell-specific glycoproteins involved in sea urchin embryonic skeleton formation. Dev. Biol. 150, 294–305 (1992)
Farach-Carson, M.C., Carson, D.D., Collier, J.L., Lennarz, W.J., Park, H.R., Wright, G.C.: A calcium-binding, asparagine-linked oligosaccharide is involved in skeleton formation in the sea urchin embryo. J. Cell Biol. 109, 1289–1299 (1989)
Park, Y., Zhang, Z., Linhardt, R.J., LeMosy, E.K.: Distinct heparan sulfate compositions in wild-type and pipe-mutant eggshell matrix. Fly (Austin). 2, 175–179 (2008)
Kamimura, K., Fujise, M., Villa, F., Izumi, S., Habuchi, H., Kimata, K., Nakato, H.: Drosophila heparan sulphate 6-O-sulphotransferase(dHS6ST) gene: structure, expression and function in the formation of the tracheal system. J. Biol. Chem. 276, 17014–17021 (2001). https://doi.org/10.1074/jbc.M011354200
Dierker, T., Shao, C., Haitina, T., Zaia, J., Hinas, A., Kjellen, L.: Nematodes join the family of chondroitin sulfate-synthesizing organisms: identification of an active chondroitin sulfotransferase in Caenorhabditis elegans. Sci. Rep. 6, 34662 (2016). https://doi.org/10.1038/srep34662
Kinnunen, T., Huang, Z., Townsend, J., Gatdula, M.M., Brown, J.R., Esko, J.D., Turnbull, J.E.: Heparan 2-O-sulfotransferase, hst-2, is essential for normal cell migration in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 102, 1507–1512 (2005). https://doi.org/10.1073/pnas.0401591102
Townley, R.A., Bulow, H.E.: Genetic analysis of the heparan modification network in Caenorhabditis elegans. J. Biol. Chem. 286, 16824–16831 (2011). https://doi.org/10.1074/jbc.M111.227926
Qian, Y., Lee, I., Lee, W.S., Qian, M., Kudo, M., Canfield, W.M., Lobel, P., Kornfeld, S.: Functions of the α, β, and γ subunits of UDP-GlcNAc:lysosomal enzyme N-acetylglucosamine-1-phosphotransferase. J. Biol. Chem. 285, 3360–3370 (2010). https://doi.org/10.1074/jbc.M109.068650
Couso, R., Lang, L., Roberts, R.M., Kornfeld, S.: Phosphorylation of the oligosaccharide of uteroferrin by UDP-GlcNAc:glycoprotein N-acetylglucosamine-1-phosphotransferases from rat liver, Acanthamoeba castellani, and Dictyostelium discoideum requires α1,2-linked mannose residues. J. Biol. Chem. 261, 6326–6331 (1986)
Qian, Y., West, C.M., Kornfeld, S.: UDP-GlcNAc:glycoprotein N-acetylglucosamine-1-phosphotransferase mediates the initial step in the formation of the methylphosphomannosyl residues on the high mannose oligosaccharides of Dictyostelium discoideum glycoproteins. Biochem. Biophys. Res. Commun. 393, 678–681 (2010)
Gabel, C.A., Costello, C.E., Reinhold, V.N., Kurz, L., Kornfeld, S.: Identification of methylphosphomannosyl residues as components of the high mannose oligosaccharides of Dictyostelium discoideum glycoproteins. J. Biol. Chem. 259, 13762–13769 (1984)
Hykollari, A., Dragosits, M., Rendic, D., Wilson, I.B., Paschinger, K.: N-glycomic profiling of a glucosidase II mutant of Dictyostelium discoideum by “off-line” liquid chromatography and mass spectrometry. Electrophoresis. 35(2116–2129), (2014). https://doi.org/10.1002/elps.201300612
Hykollari, A., Malzl, D., Yan, S., Wilson, I.B.H., Paschinger, K.: Hydrophilic interaction anion exchange for separation of multiply modified neutral and anionic Dictyostelium N-glycans. Electrophoresis. 38, 2175–2183 (2017). https://doi.org/10.1002/elps.201700073
Paschinger, K., Hykollari, A., Razzazi-Fazeli, E., Greenwell, P., Leitsch, D., Walochnik, J., Wilson, I.B.H.: The N-glycans of Trichomonas vaginalis contain variable core and antennal modifications. Glycobiology. 22, 300–313 (2012)
Hernandez, L.M., Ballou, L., Alvarado, E., Tsai, P.K., Ballou, C.E.: Structure of the phosphorylated N-linked oligosaccharides from the mnn9 and mnn10 mutants of Saccharomyces cerevisiae. J. Biol. Chem. 264, 13648–13659 (1989)
Wang, X.-H., Nakayama, K., Shimma, Y., Tanaka, A., Jigami, Y.: MNN6, a member of the KRE2/MNT1 family, is the gene for mannosylphosphate transfer in Saccharomyces cerevisiae. J. Biol. Chem. 272, 18117–18124 (1997)
Allen, S., Richardson, J.M., Mehlert, A., Ferguson, M.A.J.: Structure of a complex phosphoglycan epitope from gp72 of Trypanosoma cruzi. J. Biol. Chem. 288, 11093–11105 (2013). https://doi.org/10.1074/jbc.M113.452763
Moody-Haupt, S., Patterson, J.H., Mirelman, D., McConville, M.J.: The major surface antigens of Entamoeba histolytica trophozoites are GPI-anchored proteophosphoglycans. J. Mol. Biol. 297, 409–420 (2000)
Mehta, D.P., Ichikawa, M., Salimath, P.V., Etchison, J.R., Haak, R., Manzi, A., Freeze, H.H.: A lysosomal cysteine proteinase from Dictyostelium discoideum contains N-acetylglucosamine-1-phosphate bound to serine but not mannose-6-phosphate on N-linked oligosaccharides. J. Biol. Chem. 271, 10897–10903 (1996)
Kinoshita, T.: Biosynthesis and deficiencies of glycosylphosphatidylinositol. Proc Jpn Acad Ser B Phys Biol Sci. 90, 130–143 (2014)
Pöltl, G., Kerner, D., Paschinger, K., Wilson, I.B.H.: N-Glycans of the porcine nematode parasite Ascaris suum are modified with phosphorylcholine and core fucose residues. FEBS J. 274, 714–726 (2007). https://doi.org/10.1111/j.1742-4658.2006.05615.x
Paschinger, K., Wilson, I.B.H.: Two types of galactosylated fucose motifs are present on N-glycans of Haemonchus contortus. Glycobiology. 25, 585–590 (2015). https://doi.org/10.1093/glycob/cwv015
Jiménez-Castells, C., Vanbeselaere, J., Kohlhuber, S., Ruttkowski, B., Joachim, A., Paschinger, K.: Gender and developmental specific N-glycomes of the porcine parasite Oesophagostomum dentatum. Biochim. Biophys. Acta. 1861, 418–430 (2017). https://doi.org/10.1016/j.bbagen.2016.10.011
Yan, S., Wilson, I.B.H., Paschinger, K.: Comparison of RP-HPLC modes to analyse the N-glycome of the free-living nematode Pristionchus pacificus. Electrophoresis. 36, 1314–1329 (2015). https://doi.org/10.1002/elps.201400528
Haslam, S.M., Houston, K.M., Harnett, W., Reason, A.J., Morris, H.R., Dell, A.: Structural studies of N-glycans of filarial parasites. Conservation of phosphorylcholine-substituted glycans among species and discovery of novel chito-oligomers. J. Biol. Chem. 274, 20953–20960 (1999)
Haslam, S.M., Khoo, K.H., Houston, K.M., Harnett, W., Morris, H.R., Dell, A.: Characterisation of the phosphorylcholine-containing N-linked oligosaccharides in the excretory-secretory 62 kDa glycoprotein of Acanthocheilonema viteae. Mol. Biochem. Parasitol. 85, 53–66 (1997)
Morelle, W., Haslam, S.M., Olivier, V., Appleton, J.A., Morris, H.R., Dell, A.: Phosphorylcholine-containing N-glycans of Trichinella spiralis: identification of multiantennary lacdiNAc structures. Glycobiology. 10, 941–950 (2000)
Hanneman, A.J., Rosa, J.C., Ashline, D., Reinhold, V.: Isomer and glycomer complexities of core GlcNAcs in Caenorhabditis elegans. Glycobiology. 16, 874–890 (2006)
Paschinger, K., Hackl, M., Gutternigg, M., Kretschmer-Lubich, D., Stemmer, U., Jantsch, V., Lochnit, G., Wilson, I.B.H.: A deletion in the Golgi α-mannosidase II gene of Caenorhabditis elegans results in unexpected non-wild type N-glycan structures. J. Biol. Chem. 281, 28265–28277 (2006)
Paschinger, K., Gonzalez-Sapienza, G.G., Wilson, I.B.H.: Mass spectrometric analysis of the immunodominant glycan epitope of Echinococcus granulosus antigen Ag5. Int. J. Parasitol. 42, 279–285 (2012)
Sugita, M., Fujii, H., Dulaney, J.T., Inagaki, F., Suzuki, M., Suzuki, A., Ohta, S.: Structural elucidation of two novel amphoteric glycosphingolipids from the earthworm, Pheretima hilgendorfi. Biochim. Biophys. Acta. 1259, 220–226 (1995)
Gerdt, S., Dennis, R.D., Borgonie, G., Schnabel, R., Geyer, R.: Isolation, characterization and immunolocalization of phosphorylcholine-substituted glycolipids in developmental stages of Caenorhabditis elegans. Eur. J. Biochem. 266, 952–963 (1999)
Lochnit, G., Dennis, R.D., Ulmer, A.J., Geyer, R.: Structural elucidation and monokine-inducing activity of two biologically active zwitterionic glycosphingolipids derived from the porcine parasitic nematode Ascaris suum. J. Biol. Chem. 273, 466–474 (1998)
Noda, N., Tanaka, R., Miyahara, K., Kawasaki, T.: Isolation and characterization of a novel type of glycosphingolipid from Neanthes diversicolor. Biochim. Biophys. Acta. 1169, 30–38 (1993)
Vanbeselaere, J., Yan, S., Joachim, A., Paschinger, K., Wilson, I.B.H.: The parasitic nematode Oesophagostomum dentatum synthesizes unusual glycosaminoglycan-like O-glycans. Glycobiology. 28, 474–481 (2018). https://doi.org/10.1093/glycob/cwy045
Simenel, C., Coddeville, B., Delepierre, M., Latgé, J.P., Fontaine, T.: Glycosylinositolphosphoceramides in Aspergillus fumigatus. Glycobiology. 18, 84–96 (2008). https://doi.org/10.1093/glycob/cwm122
Unkefer, C.J., Jackson, C., Gander, J.E.: The 5-O-β-D-galactofuranosyl-containing glycopeptide from Penicillium charlesii. Identification of phosphocholine attached to C-6 of mannopyranosyl residues of the mannan region. J. Biol. Chem. 257, 2491–2497 (1982)
Maes, E., Garenaux, E., Strecker, G., Leroy, Y., Wieruszeski, J.M., Brassart, C., Guerardel, Y.: Major O-glycans from the nest of Vespula germanica contain phospho-ethanolamine. Carbohydr. Res. 340, 1852–1858 (2005). https://doi.org/10.1016/j.carres.2005.05.008
Seppo, A., Moreland, M., Schweingruber, H., Tiemeyer, M.: Zwitterionic and acidic glycosphingolipids of the Drosophila melanogaster embryo. Eur. J. Biochem. 267, 3549–3558 (2000)
Helling, F., Dennis, R.D., Weske, B., Nores, G., Peter-Katalinic, J., Dabrowski, U., Egge, H., Wiegandt, H.: Glycosphingolipids in insects. The amphoteric moiety, N-acetylglucosamine-linked phosphoethanolamine, distinguishes a group of ceramide oligosaccharides from the pupae of Calliphora vicina (Insecta: Diptera). Eur. J. Biochem. 200, 409–421 (1991)
Hykollari, A., Eckmair, B., Voglmeir, J., Jin, C., Yan, S., Vanbeselaere, J., Razzazi-Fazeli, E., Wilson, I.B., Paschinger, K.: More than just Oligomannose: an N-glycomic comparison of Penicillium species. Mol. Cell. Proteomics. 15, 73–92 (2016). https://doi.org/10.1074/mcp.M115.055061
Itasaka, O., Hori, T.: Studies on glycosphingolipids of fresh-water bivalves. V. the structure of a novel ceramide octasaccharide containing mannose-6-phosphate found in the bivalve, Corbicula sandai. J. Biochem. 85, 1469–1481 (1979)
Cipollo, J.F., Awad, A., Costello, C.E., Robbins, P.W., Hirschberg, C.B.: Biosynthesis in vitro of Caenorhabditis elegans phosphorylcholine oligosaccharides. Proc. Natl. Acad. Sci. U. S. A. 101, 3404–3408 (2004)
Houston, K.M., Sutharsan, R., Steiger, C.N., Schachter, H., Harnett, W.: Gene inactivation confirms the identity of enzymes involved in nematode phosphorylcholine-N-glycan synthesis. Mol. Biochem. Parasitol. 157, 88–91 (2008)
Ishida, N., Irikura, D., Matsuda, K., Sato, S., Sone, T., Tanaka, M., Asano, K.: Molecular cloning and expression of a novel cholinephosphotransferase involved in glycoglycerophospholipid biosynthesis of mycoplasma fermentans. Curr. Microbiol. 58, 535–540 (2009). https://doi.org/10.1007/s00284-009-9362-6
Mikolajek, H., Kolstoe, S.E., Pye, V.E., Mangione, P., Pepys, M.B., Wood, S.P.: Structural basis of ligand specificity in the human pentraxins, C-reactive protein and serum amyloid P component. J. Mol. Recognit. 24, 371–377 (2011). https://doi.org/10.1002/jmr.1090
Goodridge, H.S., Marshall, F.A., Else, K.J., Houston, K.M., Egan, C., Al-Riyami, L., Liew, F.Y., Harnett, W., Harnett, M.M.: Immunomodulation via novel use of TLR4 by the filarial nematode phosphorylcholine-containing secreted product, ES-62. J. Immunol. 174, 284–293 (2005)
Matsuura, F.: The identification of aminoalkylphosphonyl cerebrosides in the marine gastropod, Monodonta labio. J. Biochem. 85, 433–441 (1979)
Araki, S., Satake, M., Ando, S., Hayashi, A., Fujii, N.: Characterization of a diphosphonopentaosylceramide containing 3-O-methylgalactose from the skin of Aplysia kurodai (sea hare). J. Biol. Chem. 261, 5138–5144 (1986)
Araki, S., Abe, S., Ando, S., Fujii, N., Satake, M.: Isolation and characterization of a novel 2-aminoethylphosphonyl-glycosphingolipid from the sea hare, Aplysia kurodai. J. Biochem. 101, 145–152 (1987)
Araki, S., Abe, S., Odani, S., Ando, S., Fujii, N., Satake, M.: Structure of a triphosphonopentaosylceramide containing 4-O-methyl-N-acetylglucosamine from the skin of the sea hare, Aplysia kurodai. J. Biol. Chem. 262, 14141–14145 (1987)
Hayashi, A., Matsubara, T.: A new homolog of Phosphonoglycosphingolipid, N-Methylaminoethylphosphonyltrigalactosylceramide. Biochim. Biophys. Acta. 1006, 89–96 (1989). https://doi.org/10.1016/0005-2760(89)90327-5
Araki, S., Yamada, S., Abe, S., Waki, H., Kon, K., Itonori, S., Sugita, M., Ando, S.: Characterization of a novel triphosphonooctaosylceramide from the eggs of the sea hare, Aplysia kurodai. J. Biochem. 129, 93–100 (2001)
Macrae, J.I., Acosta-Serrano, A., Morrice, N.A., Mehlert, A., Ferguson, M.A.: Structural characterization of NETNES, a novel glycoconjugate in Trypanosoma cruzi epimastigotes. J. Biol. Chem. 280, 12201–12211 (2005)
Hård, K., Van Doorn, J.M., Thomas-Oates, J.E., Kamerling, J.P., Van der Horst, D.J.: Structure of the Asn-linked oligosaccharides of apolipophorin III from the insect Locusta migratoria. Carbohydrate- linked 2-aminoethylphosphonate as a constituent of a glycoprotein. Biochemistry. 32, 766–775 (1993)
Urai, M., Nakamura, T., Uzawa, J., Baba, T., Taniguchi, K., Seki, H., Ushida, K.: Structural analysis of O-glycans of mucin from jellyfish (Aurelia aurita) containing 2-aminoethylphosphonate. Carbohydr. Res. 344, 2182–2187 (2009). https://doi.org/10.1016/j.carres.2009.08.001
Acknowledgements
Open access funding provided by Austrian Science Fund (FWF). Work in our laboratory relevant to this review has been funded by the Austrian Science Fund (FWF; grants P21946 and P25058 to K.P. and P23922 and P29466 to I.B.H.W.) and the European Union (Glycopar EU FP7 Marie Curie Initial Training Network; PITN-GA-2013-608295). We thank Daniel Malzl for help in preparing figures.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Paschinger, K., Wilson, I.B.H. Anionic and zwitterionic moieties as widespread glycan modifications in non-vertebrates. Glycoconj J 37, 27–40 (2020). https://doi.org/10.1007/s10719-019-09874-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10719-019-09874-2