Abstract
While mechanistic explanation and, to a lesser extent, nomological explanation are well-explored topics in the philosophy of biology, topological explanation is not. Nor is the role of diagrams in topological explanations. These explanations do not appeal to the operation of mechanisms or laws, and extant accounts of the role of diagrams in biological science explain neither why scientists might prefer diagrammatic representations of topological information to sentential equivalents nor how such representations might facilitate important processes of explanatory reasoning unavailable to scientists who restrict themselves to sentential representations. Accordingly, relying upon a case study about immune system vulnerability to attacks on CD4+ T-cells, I argue that diagrams group together information in a way that avoids repetition in representing topological structure, facilitate identification of specific topological properties of those structures, and make available to controlled processing explanatorily salient counterfactual information about topological structures, all in ways that sentential counterparts of diagrams do not.




Similar content being viewed by others
Notes
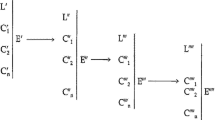
Specifically, given an abstract space E for representing relations among the parts of systems and a set of particular relations S′ in E representing the relations among the parts of a particular system S, the topological properties of S are the properties that are invariant under continuous transformations of S′ in E (Huneman 2010: 216).
According to Mach, the “goal which [physical science] has set itself is the simplest and most economical abstract expression of facts” (1895: 207; see also Duhem 1991: 21–24). Insofar as biological science aims for this economy of thought, there is an important sense in which Kitano and Oda's diagram marks scientific progress. For their diagram brings order to the facts about the immune system cellular interaction pathway, and it does so in a way superior to its sentential counterpart. I thank an anonymous referee for this observation.
The strongly connected components of a graph are its strongly connected subgraphs; a subgraph is strongly connected just in case each of its nodes is reachable from every other node; and a node w is reachable from a node v just in case there is a path from v to w, where a path from v to w is a sequence of nodes v = v o , v 1 , …, v n = w (distinct except for the possibility that v = w) such that, for each i = 0, 1, …, n, (v i , v i+1 ) is an arc (Yang et al. 2011).
I thank an anonymous referee for this observation.
Topological properties do not always carry the explanatory burden. For many mechanistic explanations, topological properties do minimal explanatory work. For example, we explain how an odorant produces an olfactory signal by appealing to the mechanism for the olfactory system. In doing so, however, we appeal primarily to specific activities and interactions among the olfactory system's components, and only secondarily (if at all) to properties of the olfactory system pathway that are invariant under continuous transformations. I thank an anonymous referee for prompting this clarification.
References
Batterman, R. W. (1992). Explanatory instability. Nous, 26, 325–348.
Batterman, R. W. (2001). The devil in the details: Asymptotic reasoning in explanation, reduction, and emergence. New York: Oxford University Press.
Bechtel, W. (2013). Understanding biological mechanisms: Using illustrations from circadian rhythm research. In K. Kampourakis (Ed.), The philosophy of biology: A companion for educators (pp. 487–510). Dordrecht: Springer.
Bechtel, W., & Abrahamsen, A. (2005). Explanation: A mechanistic alternative. Studies in History and Philosophy of Biology and Biomedical Sciences, 36, 421–441.
Bechtel, W., & Richardson, R. C. (2010). Discovering complexity: Decomposition and localization as strategies in scientific research. Cambridge, MA: MIT Press.
Broder, A., Kumar, R., Maghoul, F., Raghavan, P., Rajagopalan, S., State, R., et al. (2000). Graphical structure in the web. Computer Networks, 33, 309–320.
Carney, W. P., Rubin, R. H., Hoffman, R. A., Hansen, W. P., Healey, K., & Hirsch, M. S. (1981). Analysis of T lymphocyte subsets in cytomegalovirus mononucleosis. Journal of Immunology, 126, 2114–2116.
Cho, K.-H., & Wolkenhauer, O. (2003). Analysis and modelling of signal transduction pathways in systems biology. Biochemical Society Transactions, 31, 1503–1509.
Cook, D. L., Farley, J. F., & Tapscott, S. J. (2001). A basis for a visual language for describing, archiving and analyzing functional models of complex biological systems. Genome Biology, 2(4), research0012.2–research0012.10.
Craver, C. (2007). Constitutive explanatory relevance. Journal of Philosophical Research, 32, 3–20.
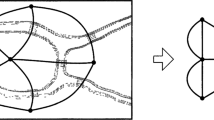
Csete, M., & Doyle, J. (1999). Bow ties, metabolism and disease. Trends in Biotechnology, 22, 446–450.
Duhem, P. (1991). The aim and structure of physical theory. Princeton: Princeton University Press.
Grossman, Z., Meier-Schellersheim, M., Sousa, A. E., Victorino, R. M. M., & Paul, W. E. (2002). CD4+ T-cell depletion in HIV infection: Are we closer to understanding the cause? Nature Medicine, 8, 319–323.
Hegarty, M. (1992). Mental animation: Inferring motion from static displays of mechanical systems. Journal of Experimental Psychology. Learning, Memory, and Cognition, 18, 1084–1102.
Huneman, P. (2010). Topological explanations and robustness in biological sciences. Synthese, 177, 213–245.
Jones, N., & Wolkenhauer, O. (2012). Diagrams as locality aids for model construction and explanation in cell biology. Biology and Philosophy, 27, 705–721.
Kitano, H. (2004). Biological robustness. Nature Reviews Genetics, 5, 826–837.
Kitano, H., & Oda, K. (2006). Robustness trade-offs and host-microbial symbiosis in the immune system. Molecular Systems Biology, 2, 1–10.
Koedinger, K. R. (1992). Emergent properties and structural constraints: Advantages of diagrammatic representations for reasoning and learning. AAAI Technical Report, SS-92–02, 151–156.
Kulpa, Z. (2009). Main problems of diagrammatic reasoning. Part I: The generalization problem. Foundations of Science, 14, 75–96.
Landais, E. X., Saulquin, & Houssaint, E. (2005). The human T cell immune response to Epstein-Barr Virus. International Journal of Developmental Biology, 49, 285–292.
Lemke, N., Herédia, F., Barcellos, C. K., dos Reis, A. N., & Mombach, J. C. M. (2004). Essentiality and damage in metabolic networks. Bioinformatics, 20, 115–119.
Lewis, M. C. (1991). Visualization and situations. In J. Barwise, M. Gawron, G. Plotkin, & S. Tutiya (Eds.), Situation theory and its applications II (pp. 553–580). CSLI Publications: Stanford, CA.
Ma, H.-W., & Zeng, A.-P. (2003). The connectivity structure, giant strong component and centrality of metabolic networks. Bioinformatics, 19, 1423–1430.
Mach, E. (1895). Popular scientific lectures. Chicago: Open Court Publishing.
Machamer, P., Darden, L., & Craver, C. F. (2000). Thinking about mechanisms. Philosophy of Science, 67, 1–25.
Mc Manus, F. (2012). Development and mechanistic explanation. Studies in History and Philosophy of Biology and Biomedical Sciences, 43, 532–541. doi:10.1016/j.shpsc.2011.12.001.
Mehta, S., & Tagore, S. (2009). Functional module analysis in metabolomics. Chokes Advances in Computational Research, 1, 1–4.
Merico, D., Gfeller, D., & Bader, G. D. (2009). How to visually interpret biological data using networks. Nature Biotechnology, 27, 921–924.
Nelson, M. D., Zhou, E., Kiontke, K., Fradin, H., Maldonado, G., Martin, D., et al. (2011). A bow-tie genetic architecture for morphogenesis suggested by a genome-wide RNAi screen in Caenorhabditis elegans. PLoS Genetics, 7(3), e1002010. doi:10.1371/journal.pgen.1002010.
Nikiforow, S., Bottomy, K., & Miller, G. (2001). CD4+ T-cell effectors inhibit Epstein-Barr virus-induced C-cell proliferation. Journal of Virology, 75, 3740–3752.
Palumbo, M. C., Colosimo, A., & Giuliani, A. (2005). Functional essentiality from topology features in metabolic networks: A case study in yeast. FEBS Letters, 579, 4642–4646.
Perini, L. (2005). Explanation in two dimensions: Diagrams and biological explanation. Biology and Philosophy, 20, 257–269.
Polouliakh, N., Nock, R., Nielsen, F., & Kitano, H. (2009). G-Protein coupled receptor signaling architecture of mammalian immune cells. PLoS ONE, 4(1), e4189. doi:10.1371/journal.pone.0004189.
Press, J. (2009). Physical explanations and biological explanations, empirical laws and a priori laws. Biology and Philosophy, 24, 359–374.
Pržulj, N., Wigle, D. A., & Jurisica, I. (2004). Functional topology in a network of protein interactions. Bioinformatics, 20, 340–348.
Rosenberg, A. (2001). How is biological explanation possible? British Journal for the Philosophy of Science, 52, 735–760.
Saraiya, P., North, C., & Duca, K. (2005). Visualizing biological pathways: Requirements analysis, systems evaluation and research agenda. Information Visualization, 4, 191–205.
Shin, S.-J. (2012). The forgotten individual: Diagrammatic reasoning in mathematics. Synthese, 186, 149–168.
Supper, J., Spangenberg, L., Planatscher, H., Dräger, A., Schröder, A., & Zell, A. (2009). BowTieBuilder: Modeling signal transduction pathways. BMC Systems Biology, 3, 67.
Thagard, P. (2003). Pathways to biomedical discovery. Philosophy of Science, 70, 235–254.
Tieri, P., Grignolio, A., Zaikin, A., Mishto, M., Remondini, D., Castellani, G. C., et al. (2010). Network, degeneracy and bow tie Integrating paradigms and architectures to grasp the complexity of the immune system. Theoretical Biology and Medical Modeling, 7, 32.
Wingate, P. J., McAulay, K. A., Anthony, I. C., & Crawford, D. H. (2009). Regulatory T cell activity in primary and persistent Epstein-Barr infection. Journal of Medical Virology, 81, 870–877.
Yang, R., Zhuhadar, L., & Nasraoui, O. (2011). Bow-tie decomposition in directed graphs. Information Fusion (FUSION), 2011 Proceedings of the 14th International Conference on, 5–8.
Yeh, I., Hanemamp, T., Tsoka, S., Karp, P. D., & Altman, R. B. (2004). Computational analysis of Plasmodium falciparum metabolism: Organizing genomic information to facilitate drug discovery. Genome Research, 14, 917–924.
Zeng, M., Smith, A. J., Wietgrefe, S. W., Southern, P. J., Schacker, T. W., Reilly, C. S., et al. (2011). Cumulative mechanisms of lymphoid tissue fibrosis and T cell depletion in HIV-1 and SIV infections. Journal of Clinical Investigation, 121, 998–1008.
Acknowledgments
I thank William Bechtel, Philippe Huneman, Daniel Pearlberg, audiences at University of Nebraska - Omaha and the Alabama Philosophical Society, as well as two anonymous referees for helpful comments on prior versions of this paper.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jones, N. Bowtie Structures, Pathway Diagrams, and Topological Explanation. Erkenn 79, 1135–1155 (2014). https://doi.org/10.1007/s10670-014-9598-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10670-014-9598-9