Abstract
Understanding how anthropogenic disturbance affects genetic diversity is essential to appropriately incorporating genetic considerations into conservation plans. Unfortunately, we rarely have information about a population’s genetic diversity before it becomes imperiled. Here we reconstruct the historic range of the naturally rare annual mustard Streptanthus glandulosus subsp. niger (Sgn) and use herbarium specimens to quantify pre-disturbance genetic diversity. We compare this to the genetic diversity in the contemporary plant populations and to plants in the seed bank. We conclude that Sgn was recently a single, panmictic population composed of orders of magnitude more plants than exist today but experienced recent and abrupt declines following housing development. Today Sgn persists as two disjunct populations, the larger of which has retained historic levels of diversity although there is a downward trend in all measures. The smaller population has lost 21–28% of the diversity that was present only 50 years ago with an Ne ~ 5–16. The contemporary populations have differentiated from each other due to drift. The seed bank contained no novel alleles and had high levels of homozygosity, indicating that it is incapable of providing genetic rescue. This novel combination of hDNA, the aboveground plant population and the seed bank can be used to design high impact conservation plans that appropriately incorporate genetic diversity for this and other imperiled species.




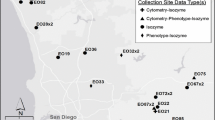
source for each sample: historic plants from the herbarium collected between 1909 and 1974, and each of the two contemporary populations (Old St. Hilary’s and Middle Ridge). OSH-SB = plants grown from the seed bank at Old St. Hilary’s; MR-SB = plants grown from the seed bank at Middle Ridge
Similar content being viewed by others
Data Availability
Data for this study will be available in the Dryad Digital Repository upon final manuscript acceptance. Prior to that, data are available from the corresponding author upon reasonable request.
Code availability
Not applicable. All data were analyzed using widely available and clearly cited freeware (GelAlEx, FSTAT, GENEPOP).
References
Aguilar R, Quesada M, Ashworth L, Herrerias-Diego Y, Lobo J (2008) Genetic consequences of habitat fragmentation in plant populations: susceptible signals in plant traits and methodological approaches. Mol Ecol 17:5177–5188
Anacker BL (2014) The nature of serpentine endemism. Am J Bot 101:219–224
Barrett SCH, Charlesworth D (1991) Effects of a change in the level of inbreeding on the genetic load. Nature 352:522–524
Berg N, Hall A (2015) Increased interannual precipitation extremes over California under climate change. J Clim 28:6324–6334
Bouzat JL, Lewin HA, Paige KN (1998) The ghost of genetic diversity past: historical DNA analysis of the greater prairie chicken. Am Nat 152:1–6
Burrell AM, Goddard JH, Greer PJ, Williams RJ, Pepper AE (2019) Sporadic genetic connectivity among small insular populations of the rare geoendemic plant Caulanthus amplexicaulis var. barbarae (Santa Barbara Jewelflower). J Hered 110:587–600
Cacho NL, Strauss SY (2014) Occupation of bare habitats, an evolutionary precursor to soil specialization in plants. Proc Natl Acad Sci 111:15132–15137
Cacho NL, Burrell AM, Pepper AE, Strauss SY (2014) Novel nuclear markers inform the systematics and the evolution of serpentine use in Streptanthus and allies (Thelypodieae, Brassicaceae). Mol Phylogenet Evol 72:71–81
California Department of Fish and Wildlife (2014) California Threatened and endangered plant profiles: Tiburon jewelflower (Streptanthus glandulosus ssp. niger). https://www.wildlife.ca.gov/Conservation/Plants/Endangered/Streptanthus-glandulosus-ssp-niger
California Native Plant Society, Rare Plant Program (2020) Inventory of Rare and Endangered Plants of California (v8-03 0.39). http://www.rareplants.cnps.org
Cohen D (1966) Optimizing reproduction in a randomly varying environment. J Theor Biol 12:119–129
Cozzolino S, Cafasso D, Pellegrino G, Musacchio A, Widmer A (2007) Genetic variation in time and space: the use of herbarium specimens to reconstruct patterns of genetic variation in the endangered orchid Anacamptis palustris. Conserv Genet 8:629–639
Deser C, Phillips AS, Alexander MA, Smoliak BV (2014) Projecting North American climate over the next 50 years: uncertainty due to internal variability. J Clim 27:2271–2296
Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR (2014) NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour 14:209–214
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Frankham R (1996) Relationship of genetic variation to population size in wildlife. Conserv Biol 10:1500–1508
Frankham R (2015) Genetic rescue of small inbred populations: meta-analysis reveals large and consistent benefits of gene flow. Mol Ecol 24(11):2610–2618
Frankham R, Ballou JD, Briscoe DA (2002) Introduction to conservation genetics. Cambridge University Press, Cambridge
Frankham R, Ballou JD, Eldridge MD, Lacy RC, Ralls K, Dudash MR, Fenster CB (2011) Predicting the probability of outbreeding depression. Conserv Biol 25:465–475
Frankham R, Bradshaw CJ, Brook BW (2014) Genetics in conservation management: revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol Conserv 170:56–63
Franklin IR (1980) Evolutionary change in small populations. In: Conservation biology: an evolutionary-ecological perspective, p 395
Garza JC, Williamson EG (2001) Detection of reduction in population size using data from microsatellite loci. Mol Ecol 10:305–318
Goudet J (1995) FSTAT Version 1.2: a computer program to calculate f-statistics. J Hered 86:485–486
Griffin D, Anchukaitis KJ (2014) How unusual is the 2012–2014 California drought? Geophys Res Lett 41:9017–9023
Hadly EA, van Tuinen M, Chan Y, Heiman K (2003) Ancient DNA evidence of prolonged population persistence with negligible genetic diversity in an endemic tuco-tuco (Ctenomys sociabilis). J Mammal 84:403–417
Hale ML, Burg TM, Steeves TE (2012) Sampling for microsatellite-based population genetic studies: 25 to 30 individuals per population is enough to accurately estimate allele frequencies. PLoS ONE 7(9):e45170
Hanin N, Quaye M, Westberg E, Barazani O (2013) Soil seed bank and among-years genetic diversity in arid populations of Eruca sativa Miller (Brassicaceae). J Arid Environ 91:151–154
Harnik PG, Simpson C, Payne JL (2012) Long-term differences in extinction risk among the seven forms of rarity. Proc R Soc B: Biol Sci 279:4969–4976
Hedrick PW, Peterson RO, Vucetich LM, Adams JR, Vucetich JA (2014) Genetic rescue in Isle Royale wolves: genetic analysis and the collapse of the population. Conserv Genet 15:1111–1121
Jorde PE, Ryman N (1995) Temporal allele frequency change and estimation of effective size in populations with overlapping generations. Genetics 139:1077–1090
Kalisz S, McPeek MA (1992) Demography of an age-structured annual: resampled projection matrices, elasticity analyses, and seed bank effects. Ecology 73:1082–1093
Kirtman B, Power SB, Adedoyin AJ, Boer GJ, Bojariu R, Camilloni I, Doblas-Reyes F, Fiore AM (2013) Chapter 11—Near-term climate change: projections and predictability. In: Climate change 2013: the physical science basis. IPCC Working Group I Contribution to AR5. Eds. IPCC. Cambridge University Press, Cambridge
Kruckeberg AR (1954) The ecology of serpentine soils III. Plant species in relation to serpentine soils. Ecology 35:267–274
Kruckeberg AR (1957) Variation in fertility of hybrids between isolated populations of the serpentine species, Streptanthus glandulosus Hook. Evolution 11:185–211
Lande R, Barrowclough G (1987) Effective population size, genetic variation, and their use in population. In: Viable populations for conservation, pp 87–123
Landguth EL, Fedy BC, Oyler-McCance SJ, Garey AL, Emel SL, Mumma M et al (2012) Effects of sample size, number of markers, and allelic richness on the detection of spatial genetic pattern. Mol Ecol Resour 12:276–284
Leck MA, Parker VT, Simpson RL (1989) Ecology of soil seed banks. Academic Press, San Diego
Lundemo S, Falahati-Anbaran M, Stenøien HK (2009) Seed banks cause elevated generation times and effective population sizes of Arabidopsis thaliana in northern Europe. Mol Ecol 18:2798–2811
Madsen T, Shine R, Olsson M, Wittzell H (1999) Restoration of an inbred adder population. Nature 402:34–35
Madsen T, Ujvari B, Olsson M (2004) Novel genes continue to enhance population growth in adders (Vipera berus). Biol Conserv 120:145–147
Mayer MS, Soltis PS, Soltis DE (1994) The evolution of the Streptanthus glandulosus complex (Cruciferae): genetic divergence and gene flow in serpentine endemics. Am J Bot 81:1288–1299
McCue KA, Holtsford TP (1998) Seed bank influences on genetic diversity in the rare annual Clarkia springvillensis (Onagraceae). Am J Bot 85:30–36
McNeely JA, Miller KR, Reid WV, Mittermeier RA, Werner TB (1990) Conserving the world's biological diversity. In: International Union for conservation of nature and natural resources
Moeller DA, Geber MA, Tiffin P (2011) Population genetics and the evolution of geographic range limits in an annual plant. Am Nat 178(S1):S44–S61
Nomura T (2008) Estimation of effective number of breeders from molecular coancestry of single cohort sample. Evol Appl 1:462–474
Noss RF (1990) Indicators for monitoring biodiversity: a hierarchical approach. Conserv Biol 4:355–364
Nunney L (2002) The effective size of annual plant populations: the interaction of a seed bank with fluctuating population size in maintaining genetic variation. Am Nat 160:195–204
Pääbo S, Poinar H, Serre D et al (2004) Genetic analyses from ancient DNA. Annu Rev Genet 38:645–679
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Resour 6:299–295
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Pickup M, Young AG (2008) Population size, self-incompatibility and genetic rescue in diploid and tetraploid races of Rutidosis leptorrhynchoides (Asteraceae). Heredity 100:268–274
Pierce DW, Das T, Cayan DR et al (2012) Probabilistic estimates of future changes in California temperature and precipitation using statistical and dynamical downscaling. Clim Dyn 40:839–856
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rabinowitz D (1981) Seven forms of rarity. In: Synge J (ed) The biological aspects of rare plant conservation. Wiley, Chichester, pp 205–217
Robinson JA, Ortega-Del Vecchyo D, Fan Z, Kim BY, Marsden CD, Lohmueller KE, Wayne RK (2016) Genomic flatlining in the endangered island fox. Curr Biol 26:1183–1189
Robinson JA, Brown C, Kim BY, Lohmueller KE, Wayne RK (2018) Purging of strongly deleterious mutations explains long-term persistence and absence of inbreeding depression in island foxes. Curr Biol 28:3487–3494
Rousset F (2008) GENEPOP ’007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
Saltonstall K (2002) Cryptic invasion by a non-native genotype of the common reed, Phragmites australis, into North America. Proc Natl Acad Sci 99:2445–2449
Särkinen T, Staats M, Richardson JE, Cowan RS, Bakker FT (2012) How to open the treasure chest? Optimising DNA extraction from herbarium specimens. PLoS ONE 7(8):e43808. https://doi.org/10.1371/journal.pone.0043808
Schwartz MK, Luikart G, Waples RS (2007) Genetic monitoring as a promising tool for conservation and management. Trends Ecol Evol 22:25–33
Sianta SA, Kay KM (2019) Adaptation and divergence in edaphic specialists and generalists: serpentine soil endemics in the California flora occur in barer serpentine habitats with lower soil calcium levels than serpentine tolerators. Am J Bot 106:690–703
Skinner MW, Pavlik BM (1994) California Native Plant Society's inventory of rare and endangered vascular plants of California. California Native Plant Society
Slatkin M (1985) Gene flow in natural populations. Annu Rev Ecol Syst 16:393–430
Slatkin M (1987) Gene flow and the geographic structure of natural populations. Science 236:787–792
Soulé ME (1980) Thresholds for survival: maintaining fitness and evolutionary potential. In: Conservation biology: an evolutionary-ecological perspective, pp 151–169
Soulé ME, Bolger DT, Alberts AC, Wrights J, Sorice M, Hill S (1988) Reconstructed dynamics of rapid extinctions of chaparral-requiring birds in urban habitat islands. Conserv Biol 2:75–92
Spurgin LG, Wright DJ, Van der Velde M, Collar NJ, Komdeur J, Burke T, Richardson DS (2014) Museum DNA reveals the demographic history of the endangered Seychelles warbler. Evol Appl 7:1134–1143
Stöcklin J, Fischer M (1999) Plants with longer-lived seeds have lower local extinction rates in grassland remnants 1950–1985. Oecologia 120:539–543
Swope SM, Pepper AE, Lee GT, Burnett BA, Horten HM (2019) Development of 15 microsatellite loci in the endangered Streptanthus glandulosus subsp. niger (Brassicaceae). Appl Plant Sci 7:1215
Templeton AR, Read B (1984) Factors eliminating inbreeding depression in a captive herd of Speke’s gazelle (Gazella spekei). Zoo Biol 3:177–199
Tracy LN, Jamieson IG (2011) Historic DNA reveals contemporary population structure results from anthropogenic effects, not pre-fragmentation patterns. Conserv Genet 12:517–526
U.S. Fish and Wildlife Service (2019) Endangered species database. www.fws.gov/endangered/species/us-species.html
Van Oosterhout C, Hutchinson WF, Wills DP, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Vandepitte K, De Meyer T, Helsen K, Van Acker K, Roldán-Ruiz I, Mergeay J, Honnay O (2014) Rapid genetic adaptation precedes the spread of an exotic plant species. Mol Ecol 23:2157–2164
Vitalis R, Glémin S, Olivieri I (2004) When genes go to sleep: the population genetic consequences of seed dormancy and monocarpic perenniality. Am Nat 163:295–311
Wandeler P, Hoeck PE, Keller LF (2007) Back to the future: museum specimens in population genetics. Trends Ecol Evol 22:634–642
Wang J (2017) The computer program structure for assigning individuals to populations: easy to use but easier to misuse. Mol Ecol Resour 17:981–990
Waples RS, Do CHI (2008) LDNE: a program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Resour 8:753–756
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Acknowledgements
We are especially grateful to the following herbaria for allowing us to destructively sample specimens: University and Jepson Herbaria (University of California, Berkeley), Hoover Herbarium (California Polytechnic State University, San Luis Obispo), J.M. Tucker and Beecher Crampton Herbaria (University of California, Davis), Southwest Environmental Information Network (Arizona), Harvard University Herbaria (Harvard University, Cambridge), Rancho Santa Ana Botanic Garden (Claremont, California), and the California Academy of Sciences Herbarium (San Francisco). Two anonymous reviewers offered thoughtful comments that improved this manuscript. Hannah Horten and Brittany Burnett assisted with lab work. We thank Dave Gotz and the Tiburon Landmark Society for access to historic photos. We thank Sam Abercrombie for creating the maps. This work was supported by a grant from Marin County Parks (14-6300-62) to SMS and Jill Barrett Foundation fellowships to TYS and NRKA. This work was conducted under a permit from the California Department of Fish and Wildlife (permit SEMP 2081(a)-14-004-RP) issued to SMS.
Funding
This work was supported by a grant from Marin County Parks (CA, USA) (award #: 14-6300-62) to SMS and Jill Barrett Foundation fellowships (through the Mills College Biology Dept.) to TYS and NRKA.
Author information
Authors and Affiliations
Contributions
SMS conceived of the study, collected leaf tissue in the field, acquired herbarium specimens and historical photographs, scored PCR results, analyzed data, conducted the literature search, wrote the manuscript. TYS and NRKA conducted the lab work, provided comments on drafts of the manuscripts, assisted with the literature search.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Ethics approval
Ethics approval not required. This work was conducted under a permit from the California Department of Fish and Wildlife (permit SEMP 2081(a)-14-004-RP) issued to SMS.
Consent to participate
Not applicable.
Consent for publication
All authors and relevant institutions (Mills College, Marin County Parks) give consent for publication of this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Swope, S.M., Soto, T.Y. & Rahman-Khan Arana, N. Historic DNA reveals genetic consequences of fragmentation in an endangered, endemic mustard. Conserv Genet 23, 123–137 (2022). https://doi.org/10.1007/s10592-021-01406-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-021-01406-6