Abstract
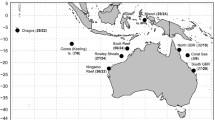
Oyster reef habitats are critical to coastal biodiversity and their decline has prompted restoration efforts in Australia. Knowledge gaps exist regarding the population structure and diversity of key species in these habitats. This may be critical information for the design of effective restoration programs. Sydney rock oysters (Saccostrea glomerata) are the dominant reef-forming bivalve in eastern Australia. Wild populations of S. glomerata have declined due to overharvesting, disease outbreaks, coastal development and reduced water quality. Here, we use genetic markers identified by genome-wide sequencing to investigate the genetic structure and diversity of wild Sydney rock oysters throughout their distribution in eastern Australia. We examine evidence for past population bottlenecks and spatial genetic structure associated with the East Australian Current. Analysis of 3, 400 neutral single-nucleotide polymorphisms (SNPs) revealed a single population, and an overlap with two other Saccostrea sp. at the northernmost boundary of the distribution. We detected signals of asymmetric gene flow consistent with the direction of the East Australian Current, and spatial structure patterns of limited genetic isolation by distance and spatial autocorrelation in the northern region (which experiences stronger effects of the East Australian Current) but not in the southern region of the distribution. We found no evidence of significant recent bottlenecks, with high effective population size throughout the species’ range. This information will provide a baseline against which to assess the impact of restoration projects, and guide strategies for sourcing stock for the enhancement of wild oyster populations. Our results provide a positive outlook for the resilience and adaptive capacity of Sydney rock oysters, and highlight wild populations as valuable resources for aquaculture and restoration initiatives.
Similar content being viewed by others
Data availability
Datasets generated and/or analysed during this project are available on Zenodo (https://doi.org/10.5281/zenodo.4589782).
Code availability
Codes for the analyses described herein are available on Zenodo (https://doi.org/10.5281/zenodo.4589782).
References
Archer FI, Adams PE, Schneiders BB (2017) strataG: An R package for manipulating, summarizing and analysing population genetic data. Mol Ecol Resour 17:5–11
Banks SC, Piggott MP, Williamson JE, Bové U, Holbrook NJ, Beheregaray LB (2007) Oceanic variability and coastal topography shape genetic structure in a long-dispersing sea urchin. Ecology 88:3055–3064
Bartoń K (2019) MuMIn: multi-model inference. R package version 1.43.6
Bateman DC, Bishop MJ (2017) The environmental context and traits of habitat-forming bivalves influence the magnitude of their ecosystem engineering. Mar Ecol Prog Ser 563:95–110
Beck MW et al (2011) Oyster reefs at risk and recommendations for conservation, restoration, and management. Bioscience 61:107–116
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B 57:289–300
Borsje BW, van Wesenbeeck BK, Dekker F, Paalvast P, Bouma TJ, van Katwijk MM, de Vries MB (2011) How ecological engineering can serve in coastal protection. Ecol Eng 37:113–122
Brook BW, Bradshaw CJA, Traill LW, Frankham R (2011) Minimum viable population size: not magic, but necessary. Trends Ecol Evol 26:619–620. https://doi.org/10.1016/j.tree.2011.09.006
Buroker NE, Hershberger W, Chew K (1979a) Population genetics of the family Ostreidae. I. Intraspecific studies of Crassostrea gigas and Saccostrea commercialis. Mar Biol 54:157–169
Buroker NE, Hershberger WK, Chew KK (1979b) Population genetics of the family Ostreidae. II. Interspecific studies of the genera Crassostrea and Saccostrea. Mar Biol 54:171–184. https://doi.org/10.1007/bf00386596
Butt D, Raftos D (2007) Immunosuppression in Sydney rock oysters (Saccostrea glomerata) and QX disease in the Hawkesbury River, Sydney. Mar Freshwater Res 58:213–221. https://doi.org/10.1071/MF06080
Camara MD, Vadopalas B (2009) Genetic aspects of restoring Olympia oysters and other native bivalves: balancing the need for action, good intentions, and the risks of making things worse. J Shellfish Res 28:121–145
Campbell CD et al (2012) Estimating the human mutation rate using autozygosity in a founder population. Nat Genet 44:1277–1281
Cavalli-Sforza LL, Edwards AW (1967) Phylogenetic analysis: models and estimation procedures. Evolution 21:550–570
Chen J-h et al (2019) Genome-wide analysis of Cushion willow provides insights into alpine plant divergence in a biodiversity hotspot. Nat Commun 10:5230. https://doi.org/10.1038/s41467-019-13128-y
Clarke RT, Rothery P, Raybould AF (2002) Confidence limits for regression relationships between distance matrices: estimating gene flow with distance. J Agr Biol Envir St 7:361
Coen LD et al (2007) Ecosystem services related to oyster restoration. Mar Ecol Prog Ser 341:303–307
Coffman AJ, Hsieh PH, Gravel S, Gutenkunst RN (2016) Computationally efficient composite likelihood statistics for demographic inference. Mol Biol Eovl 33:591–593
Coleman MA, Feng M, Roughan M, Cetina-Heredia P, Connell SD (2013) Temperate shelf water dispersal by Australian boundary currents: Implications for population connectivity. Limnol Oceanogr Fluids Environ 3:295–309. https://doi.org/10.1215/21573689-2409306
Curley BG, Gillings MR (2009) Population connectivity in the temperate damselfish Parma microlepis: analyses of genetic structure across multiple spatial scales. Mar Biol 156:381–393. https://doi.org/10.1007/s00227-008-1090-0
Danecek P et al (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158
Diggles B (2013) Historical epidemiology indicates water quality decline drives loss of oyster (Saccostrea glomerata) reefs in Moreton Bay, Australia. New Zeal J Mar Fresh 47:561–581
Dinamani P (1974) Reproductive cycle and gonadial changes in the New Zealand rock oyster Crassostrea glomerata. New Zeal J Mar Fresh 8:39–65
do Amaral VS, Simone LRL, (2016) Comparative anatomy of five species of Saccostrea Dollfus and Dautzenberg, 1920 (Bivalvia: Ostreidae) from the Pacific Ocean. Nautilus 130:53–71
Do C, Waples RS, Peel D, Macbeth G, Tillett BJ, Ovenden JR (2014) NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour 14:209–214
Dove MC, O’Connor WA (2007) Salinity and temperature tolerance of Sydney rock oysters Saccostrea glomerata during early ontogeny. J Shellfish Res 26:939–947
Edwards AWF (1971) Distances between populations on the basis of gene frequencies. Biometrics 27:873–881
Everett MV, Park LK, Berntson EA, Elz AE, Whitmire CE, Keller AA, Clarke ME (2016) Large-scale genotyping-by-sequencing indicates high levels of gene flow in the deep-sea octocoral Swiftia simplex (Nutting 1909) on the west coast of the United States. PLoS ONE 11:e0165279
Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using Phred I Accuracy assessment. Genome Res 8:175–185
Excoffier L, Lischer HE (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Foll M (2012) BayeScan v2. 1 user manual. Ecology 20:1450–1462
Foll M, Gaggiotti O (2008) A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180:977–993
Frankham R (1995) Effective population size/adult population size ratios in wildlife: a review. Genet Res 66:95–107. https://doi.org/10.1017/s0016672300034455
Frankham R, Ballou JD, Eldridge MDB, Lacy RC, Ralls K, Dudash MR, Fenster CB (2011) Predicting the probability of outbreeding depression. Conserv Biol 25:465–475. https://doi.org/10.1111/j.1523-1739.2011.01662.x
Frankham R, Lees K, Montgomery ME, England PR, Lowe EH, Briscoe DA (1999) Do population size bottlenecks reduce evolutionary potential? Anim Conserv 2:255–260. https://doi.org/10.1111/j.1469-1795.1999.tb00071.x
Franklin IR, Frankham R (1998) How large must populations be to retain evolutionary potential? Anim Conserv 1:69–70. https://doi.org/10.1111/j.1469-1795.1998.tb00228.x
Gillies CL et al (2015) Scaling-up marine restoration efforts in Australia. Ecol Manag Restor 16:84–85
Gillies CL et al (2018) Australian shellfish ecosystems: Past distribution, current status and future direction. PLoS ONE 13:e0190914
Goncalves P, Jones DB, Thompson EL, Parker LM, Ross PM, Raftos DA (2017) Transcriptomic profiling of adaptive responses to ocean acidification. Mol Ecol 26:5974–5988
Guo X, Ford SE (2016) Infectious diseases of marine molluscs and host responses as revealed by genomic tools. Philos Trans R Soc B 371:20150206
Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD (2009) Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet 5:e1000695
Hare MP, Avise JC (1996) Molecular genetic analysis of a stepped multilocus cline in the American oyster (Crassostrea virginica). Evolution 50:2305–2315. https://doi.org/10.1111/j.1558-5646.1996.tb03618.x
Hare MP et al (2011) Understanding and estimating effective population size for practical application in marine species management. Conserv Biol 25:438–449
Hoey JA, Pinsky ML (2018) Genomic signatures of environmental selection despite near-panmixia in summer flounder. Evol Appl 11:1732–1747. https://doi.org/10.1111/eva.12676
Horne JB, Momigliano P, Welch DJ, Newman SJ, Van Herwerden L (2011) Limited ecological population connectivity suggests low demands on self-recruitment in a tropical inshore marine fish (Eleutheronema tetradactylum: Polynemidae). Mol Ecol 20:2291–2306
Hsiao S-T, Chuang S-C, Chen K-S, Ho P-H, Wu C-L, Chen CA (2016) DNA barcoding reveals that the common cupped oyster in Taiwan is the Portuguese oyster Crassostrea angulata (Ostreoida; Ostreidae), not C. gigas. Sci Rep 6:34057. https://doi.org/10.1038/srep34057
Huang Q, Baum L, Fu W-L (2010) Simple and practical staining of DNA with GelRed in agarose gel electrophoresis. Clin Lab 56:149
Hurwood DA, Heasman MP, Mather PB (2005) Gene flow, colonisation and demographic history of the flat oyster Ostrea angasi. Mar Freshwater Res 56:1099–1106. https://doi.org/10.1071/MF04261
Jefferson R (2019) Aquaculture Production Report 2017-2018. NSW Department of Primary Industries, Port Stephens Fisheries Institute
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jouganous J, Long W, Ragsdale AP, Gravel S (2017) Inferring the joint demographic history of multiple populations: beyond the diffusion approximation. Genetics 206:1549–1567
Kawamura K, Miyake T, Obata M, Aoki H, Komaru A (2017) Population demography and genetic characteristics of the Pacific Oyster Crassostrea gigas in Japan. Biochem Syst Ecol 70:211–221. https://doi.org/10.1016/j.bse.2016.12.006
Keenan K, McGinnity P, Cross TF, Crozier WW, Prodöhl PA (2013) diveRsity: an R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol Evol 4:782–788
Kilian A et al. (2012) Diversity arrays technology: a generic genome profiling technology on open platforms. In: Data production and analysis in population genomics. Springer, pp 67–89
King KC, Lively CM (2012) Does genetic diversity limit disease spread in natural host populations? Heredity 109:199–203. https://doi.org/10.1038/hdy.2012.33
Korneliussen TS, Albrechtsen A, Nielsen R (2014) ANGSD: analysis of next generation sequencing data. BMC Bioinform 15:356. https://doi.org/10.1186/s12859-014-0356-4
Lal MM, Southgate PC, Jerry DR, Bosserelle C, Zenger KR (2017) Swept away: ocean currents and seascape features influence genetic structure across the 18000 Km Indo-Pacific distribution of a marine invertebrate, the black-lip pearl oyster Pinctada margaritifera. https://doi.org/10.1186/s12864-016-3410-y
Lam K, Morton B (2006) Morphological and mitochondrial-DNA analysis of the Indo-West Pacific rock oysters (Ostreidae: Saccostrea species). J Molluscan Stud 72:235–245. https://doi.org/10.1093/mollus/eyl002
Langella O, Chikhi L, Beaumont MA (2001) lea (likelihood-based estimation of admixture): a program to estimate simultaneously admixture and time since the admixture event. Mol Ecol Notes 1:357–358. https://doi.org/10.1046/j.1471-8278.2001.00099.x
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357
Lazoski C, Gusmão J, Boudry P, Solé-Cava AM (2011) Phylogeny and phylogeography of Atlantic oyster species: evolutionary history, limited genetic connectivity and isolation by distance. Mar Ecol Prog Ser 426:197–212
Lenihan HS, Micheli F, Shelton SW, Peterson CH (1999) The influence of multiple environmental stressors on susceptibility to parasites: an experimental determination with oysters. Limnol Oceanogr 44:910–924
Leung TLF, Bates AE (2013) More rapid and severe disease outbreaks for aquaculture at the tropics: implications for food security. J Appl Ecol 50:215–222. https://doi.org/10.1111/1365-2644.12017
Li H et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Lischer H, Excoffier L (2012) PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics 28:298–299
Liu X (2020) Human prehistoric demography revealed by the polymorphic pattern of CpG transitions. Mol Biol Evol. https://doi.org/10.1093/molbev/msaa112
Liu X, Fu Y-X (2015) Exploring population size changes using SNP frequency spectra. Nat Genet 47:555. https://doi.org/10.1038/ng.3254
Lotterhos KE, Whitlock MC (2014) Evaluation of demographic history and neutral parameterization on the performance of FST outlier tests. Mol Ecol 23:2178–2192
Lynch M, Gutenkunst R, Ackerman M, Spitze K, Ye Z, Maruki T, Jia Z (2017) Population genomics of Daphnia pulex. Genetics 206:315. https://doi.org/10.1534/genetics.116.190611
Lynch M, Lande R (1998) The critical effective size for a genetically secure population. Anim Conserv 1:70–72. https://doi.org/10.1111/j.1469-1795.1998.tb00229.x
Mackenzie CL, Lynch SA, Culloty SC, Malham SK (2014) Future oceanic warming and acidification alter immune response and disease status in a commercial shellfish species, Mytilus edulis L. PLoS ONE 9:e99712–e99712. https://doi.org/10.1371/journal.pone.0099712
McAfee D, Bishop MJ, Yu TN, Williams GA (2018) Structural traits dictate abiotic stress amelioration by intertidal oysters. Funct Ecol 32:2666–2677
McKenna A et al (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
McLeod I, Schmider J, Creighton C, Gillies C (2018) Seven pearls of wisdom: advice from Traditional Owners to improve engagement of local Indigenous people in shellfish ecosystem restoration. Ecol Manag Restor 19:98–101
McLeod IM, zu Ermgassen PS, Gillies CL, Hancock B, Humphries A (2019) Can Bivalve Habitat Restoration Improve Degraded Estuaries? In: Coasts and Estuaries. Elsevier, pp 427–442
Melwani AR (2016) The biology of environmental stress: adaptive responses in Sydney rock oysters (Saccostrea glomerata) from an urbanized estuary. Dissertation, Macquarie University
Momigliano P, Harcourt R, Robbins W, Jaiteh V, Mahardika G, Sembiring A, Stow A (2017) Genetic structure and signatures of selection in grey reef sharks (Carcharhinus amblyrhynchos). Heredity 119:142–153
Momigliano P, Florin A-B, Merilä J (2021) Biases in demographic modelling affect our understanding of recent divergence. Mol Biol Evol. https://doi.org/10.1093/molbev/msab047
Nayfa MG, Zenger KR (2016) Unravelling the effects of gene flow and selection in highly connected populations of the silver-lip pearl oyster (Pinctada maxima). Mar Genom 28:99–106
Nell JA (1993) Farming the Sydney rock oyster (Saccostrea commercialis) in Australia. Rev Fish Sci 1:97–120
Nell JA (2001) The history of oyster farming in Australia. Mar Fish Rev 63:14–25
Nell JA, Smith IR, McPhee CC (2000) The Sydney rock oyster Saccostrea glomerata (Gould 1850) breeding programme: progress and goals. Aquac Res 31:45–49
Newell RI (2004) Ecosystem influences of natural and cultivated populations of suspension-feeding bivalve molluscs: a review. J Shellfish Res 23:51–62
Ogburn DM, White I, McPhee DP (2007) The disappearance of oyster reefs from Eastern Australian Estuaries: impact of colonial settlement or Mudworm invasion? Coast Manage 35:271–287. https://doi.org/10.1080/08920750601169618
Olivier F, Ridd M, Klumpp D (2002) The use of transplanted cultured tropical oysters (Saccostrea commercialis) to monitor Cd levels in North Queensland coastal waters (Australia). Mar Pollut Bull 44:1051–1062. https://doi.org/10.1016/s0025-326x(02)00157-1
Patton AH et al (2019) Contemporary demographic reconstruction methods are robust to genome assembly quality: a case study in Tasmanian Devils. Mol Biol Evol 36:2906–2921
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Pereira RR, Scanes E, Parker LM, Byrne M, Cole VJ, Ross PM (2019) Restoring the flat oyster Ostrea angasi in the face of a changing climate. Mar Ecol Prog Ser 625:27–39
Pérez-Portela R et al (2018) Genetic homogeneity of the invasive lionfish across the Northwestern Atlantic and the Gulf of Mexico based on Single Nucleotide Polymorphisms. Sci Rep 8:5062. https://doi.org/10.1038/s41598-018-23339-w
Pinheiro J, Bates D, DebRoy S, Sarkar D, R Core Team (2012) nlme: Linear and nonlinear mixed effects models. R package version 3
Pinsky ML, Palumbi SR (2014) Meta-analysis reveals lower genetic diversity in overfished populations. Mol Ecol 23:29–39. https://doi.org/10.1111/mec.12509
Plough LV (2016) Genetic load in marine animals: a review. Curr Zool 62:567–579
Portik DM et al (2017) Evaluating mechanisms of diversification in a Guineo-Congolian tropical forest frog using demographic model selection. Mol Ecol 26:5245–5263
Powell D, Subramanian S, Suwansa-ard S, Zhao M, O’Connor W, Raftos D, Elizur A (2018) The genome of the oyster Saccostrea offers insight into the environmental resilience of bivalves. DNA Res 25:655–665. https://doi.org/10.1093/dnares/dsy032
R Core Team (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Reed DH, Frankham R (2003) Correlation between fitness and genetic diversity. Conserv Biol 17:230–237. https://doi.org/10.1046/j.1523-1739.2003.01236.x
Rey Benayas JM, Newton AC, Diaz A, Bullock JM (2009) Enhancement of biodiversity and ecosystem services by ecological restoration: a meta-analysis. Science 325:1121–1124
Rose CG, Paynter KT, Hare MP (2006) Isolation by distance in the Eastern Oyster, Crassostrea virginica, in Chesapeake Bay. J Hered 97:158–170. https://doi.org/10.1093/jhered/esj019
Silliman K (2019) Population structure, genetic connectivity, and adaptation in the Olympia oyster (Ostrea lurida) along the west coast of North America. Evol Appl 12:923–939. https://doi.org/10.1111/eva.12766
Spielman D, Brook BW, Briscoe DA, Frankham R (2004) Does inbreeding and loss of genetic diversity decrease disease resistance? Conserv Genet 5:439–448. https://doi.org/10.1023/B:COGE.0000041030.76598.cd
Summerhayes SA, Kelaher BP, Bishop MJ (2009) Spatial patterns of wild oysters in the Hawkesbury River, NSW, Australia. J Shellfish Res 28:447–452
Sumpton WD, Ovenden JR, Keenan CP, Street R (2008) Evidence for a stock discontinuity of snapper (Pagrus auratus) on the east coast of Australia. Fish Res 94:92–98. https://doi.org/10.1016/j.fishres.2008.07.001
Tan K, Zhang H, Zheng H (2020) Selective breeding of edible bivalves and its implication of global climate change. Rev Aquacult. https://doi.org/10.1111/raq.12458
Tan K, Zheng H (2019) Climate change and bivalve mass mortality in temperate regions. In: de Voogt P (ed) Reviews of environmental contamination and toxicology, vol 251. Springer, Cham, pp 109–129
Teske PR, Sandoval-Castillo J, van Sebille E, Waters J, Beheregaray LB (2015) On-shelf larval retention limits population connectivity in a coastal broadcast spawner. Mar Ecol Prog Ser 532:1–12
Thomson J (1954) The genera of oysters and the Australian species. Mar Freshwater Res 5:132–168
Vignal A, Milan D, SanCristobal M, Eggen A (2002) A review on SNP and other types of molecular markers and their use in animal genetics. Genet Sel Evol 34:1
Wall CC, Peterson BJ, Gobler CJ (2008) Facilitation of seagrass Zostera marina productivity by suspension-feeding bivalves. Mar Ecol Prog Ser 357:165–174
Whitlock MC, Lotterhos KE (2015) Reliable detection of loci responsible for local adaptation: inference of a null model through trimming the distribution of FST. Am Nat 186:S24–S36. https://doi.org/10.1086/682949
Wickham H (2016) ggplot2: elegant graphics for data analysis. Springer, New York
Wijeratne S, Pattiaratchi C, Proctor R (2018) Estimates of surface and subsurface boundary current transport around Australia. J Geophys Res Oceans 123:3444–3466
Zannella C et al (2017) Microbial diseases of bivalve Mollusks: infections, immunology and antimicrobial defense. Mar Drugs s 15:182. https://doi.org/10.3390/md15060182
Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS (2012) A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28:3326–3328
Acknowledgements
The authors extend sincere thanks to the oyster industry in eastern Australia for sharing knowledge of local oyster populations, and Shana Ahmed, Liam O’Hare, Cathy James, Peter James and Maria Vozzo for assistance with fieldwork. This work was funded in part by an Australian Research Council Discovery Grant #DP150101363 to DAR (with Melanie J. Bishop), an Australian Research Council Industrial Transformation Training Centre Grant #IC130100009 to DAR (with Paul A. Haynes, Wayne A. O'Connor and others), and Macquarie University. Two anonymous reviewers are thanked for their insightful and constructive comments on an earlier version of this manuscript.
Funding
This work was funded in part by an Australian Research Council Discovery Grant #DP150101363 to DAR (with Melanie J. Bishop), an Australian Research Council Industrial Transformation Training Centre Grant #IC130100009 to DAR (with Paul A. Haynes, Wayne A. O'Connor and others), and Macquarie University.
Author information
Authors and Affiliations
Contributions
JAO, DAR and AJS conceived the project; JAO collected the samples and generated data; DAR, AJS and PM supervised the research; JAO, DAR, AJS and PM developed or designed methods; JAO analysed the data; JAO wrote the paper; DAR, AJS and PM substantially edited the paper; PM contributed substantial materials and resources.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethics approval
Sampling was conducted under General Fisheries Permit 188883 (Queensland), Scientific Collection Permit P07/0047-6.0 (New South Wales), Fisheries General Research Permit RP1288 (Victoria) and National Parks Research Permit 10008300 (Croajingolong National Park). Sample processing at Macquarie University was completed under Biosafety Permit 5201600675.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
O’Hare, J.A., Momigliano, P., Raftos, D.A. et al. Genetic structure and effective population size of Sydney rock oysters in eastern Australia. Conserv Genet 22, 427–442 (2021). https://doi.org/10.1007/s10592-021-01343-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-021-01343-4