Abstract
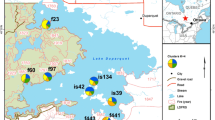
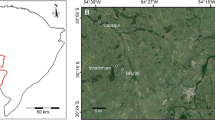
Understanding patterns of genetic diversity at the landscape scale will enhance conservation and management of natural populations. Here we analyzed the genetic diversity, population connectivity, and spatial genetic structure among subpopulations and age groups of Olea europaea subsp. cuspidata, a cornerstone species of the Afromontane highlands. The study was conducted at the landscape level within a radius of approximately 4 km, as well as on a fine scale (intensive study plot) of less than 300 m radius. In total 542 samples from four natural subpopulations in northwestern Ethiopia were analyzed using ten nuclear microsatellite markers. Inbreeding was higher in smaller populations. No genetic difference was detected among cohorts of different tree sizes in the intensive studied plot. Average population differentiation was low but significant (F ST = 0.016). Landscape genetic analysis inferred two groups: the most distant subpopulation WE located less than 4 kms from the other three subpopulations formed a separate group. Sixty-four percent of the total migrants were shared among the three latter subpopulations, which are spatially clustered. Immigrants were non-randomly distributed inside of the intensive study plot. Significant spatial genetic structure (SGS) was found both at the landscape scale and in the intensive study plot, and adults showed stronger SGS than young trees. An indirect estimate of 220 m as mean gene dispersal distance was obtained. We conclude that even under fragmentation migration is not disrupted in wild olive trees and that large protected populations at church forests are very important to conserve genetic resources. However, the higher level of inbreeding and evidence for population bottlenecks in the small populations, as well as the persisting heavy pressure on most remaining populations, warrants quick action to maintain genetic diversity of wild olive in the Ethiopian highlands.





Similar content being viewed by others
References
Abiyu HA (2012) The role of seed dispersal, exclosures, nurse shrubs and trees around churches and farms for restoration of ecosystem diversity and productivity in the Ethiopian highlands. Dissertation, University of Natural Resources and Life sciences, Vienna
Abiyu A, Teketay D, Glatzel G, Gratzer G (2015) Tree seed dispersal by African civets in the Afromontane highlands: too long a latrine to be effective for tree population dynamics. Afr J Ecol 53:588–591
Aerts R, Van Overtveld K, November E, Wassie A, Abiyu A, Demissew S, Daye DD, Giday K, Haile M, TewoldeBerhan S, Teketay D, Teklehaimanot Z, Binggeli P, Deckers J, Friis I, Gratzer G, Hermy M, Heyn M, Honnay O, Paris M, Sterck FJ, Muys B, Bongers F, Healey JR (2016) Conservation of the Ethiopian church forests: threats, opportunities and implications for their management. Sci Total Environ. doi:10.1016/j.scitotenv.2016.02.034
Anderson CD, Epperson BK, Fortin MJ, Holderegger R, James PMA, Rosenberg MS, Scribner KT, Spear S (2010) Considering spatial and temporal scale in landscape-genetic studies of gene flow. Mol Ecol 19:3565–3575
Baali-Cherif D, Besnard G (2005) High genetic diversity and clonal growth in relict populations of Olea europaea subsp. laperrinei (Oleaceae) from Hoggar, Algeria. Ann Bot 96:823–830
Balkenhol N, Cushman S, Storfer A, Waits L (2016) Landscape genetics: concepts, methods, applications, 1st edn. Wiley Blackwell, London
Bekele T (2005) Recruitment, survival and growth of Olea europeaea subsp. cuspidata seedlings and juveniles in dry Afromontane forests of northern Ethiopia. Trop Ecol 46:113–126
Besag J (1977) Contribution to the discussion of Dr. Replay’s paper. J Roy Stat Soc B 39:193–195
Besnard G, Khadari B, Villemur P, Bervillé A (2000) Cytoplasmic male sterility in the olive (Olea europaea L.). Theor Appl Genet 100:1018–1024
Besnard G, Christin PA, Baali-Cherif D, Bouguedoura N, Anthelme F (2007) Spatial genetic structure in the Laperrinei’s olive (Olea europaea subsp. laperrinei), a long living tree from the central Saharan mountains. Heredity 99:649–657
Besnard G, García-Verdugo C, Rubio De Casas R, Treier UA, Galland N, Vargas P (2008) Polyploidy in the olive complex (Olea europaea): evidence from flow cytometry and nuclear microsatellite analyses. Ann Bot 101:25–30
Besnard G, Baali-Cherif D, Bettinelli-Riccardi S, Bouguedoura N (2009) Pollen-mediated gene flow in a highly fragmented landscape: consequences for defining a conservation strategy of the relict Laperrine’s olive. CR Biol 332:662–672
Besnard G, El Bakkali A, Haouane H, Baali-Cherif D, Moukhli A, Khadari B (2013) Population genetics of Mediterranean and Saharan olives: geographic patterns of differentiation and evidence for early generation of admixture. Ann Bot 112:1293–1302
Breton C, Bervillé A (2013) From the olive flower to the drupe: flower types, pollination, self and inter-compatibility and fruit set. In: Barbara S (ed) The Mediterranean genetic code—grapevine and olive, In Tech, pp 291–213
Breton C, Tersac M, Bervillé A (2006) Genetic diversity and gene flow between the wild olive (oleaster, Olea europaea L.) and the olive: several Plio-Pleistocene refuge zones in the Mediterranean basin suggested by simple sequence repeats analysis. J Biogeogr 33:1916–1928
Breton, CM, Farinelli D, Shafiq S, Heslop-Harrison JS, Sedgley M, Bervillé AJ (2014) The self-incompatibility mating system of the olive (Olea europaea L.) functions with dominance between S- alleles. Tree Genet Genom 10:1055–1067
Carriero F, Fontanazza G, Cellini F, Giorio G (2002) Identification of simple sequence repeats (microsatellites) in olive (Olea europaea L.). Theor Appl Genet 104:301–307
Chybicki IJ, Burczyk J (2009) Simultaneous estimation of null alleles and inbreeding coefficients. J Hered 100:106–113
Cipriani G, Marrazzo MT, Marconi R, Cimato A, Testolin R (2002) Microsatellite markers isolated in olive (Olea europaea L.) are suitable for individual fingerprinting and reveal polymorphism within ancient cultivars. Theor Appl Genet 104:223–228
Collevatti RG, Estolano R, Rebeiro ML, Rabelo SG, Lima EJ, Munhoz CBR (2014) High genetic diversity and contrasting fine-scale spatial genetic structure in four seasonally dry tropical forest tree species. Plant Syst Evol 300:1671–1681
Cruse-Sanders JM, Hamrick JL (2004) Spatial and genetic structure within populations of wild American ginseng (Panax quinquefolius L., Araliaceae). J Hered 95:309–321
Cuneo P, Leishman MR (2006) African olive (Olea europeaea subsp. cuspidata) as an environmental weed in Eastern Australia: a review. Cunninghamia 9:545–577
Darbyshire I, Lamb H, Umer M (2003) Forest clearance and regrowth in northern Ethiopia during the last 3000 years. Holocene 13:537–546
De La Rosa R, James CM, Tobutt KR (2002) Isolation and characterization of polymorphic microsatellites in olive (Olea europaea L.) and their transferability to other genera in the Oleaceae. Mol Ecol Notes 2:265–267
Desalegn G, Tadesse W (2010) Major characteristics and potential uses of Eucalyptus timber species grown in Ethiopia. In: Gil L, Tadesse W, Tolosana E, López R (eds) Eucalyptus species management, history, status and trends in Ethiopia. Proceedings of the congress, Ethiopian institute of agricultural research, Addis Ababa, pp 29–52
Dias PC (1996) Sources and sinks in population biology. Trends Ecol Evol 11:326–330
Doligez A, Baril C, Joly HI (1998) Fine-scale spatial genetic structure with nonuniform distribution of individuals. Genetics 148:905–919
Escobar-Páramo P (2000) Microsatellite primers for the wild brown capuchin monkey Cebus apella. Mol Ecol 9:107–118
Fuchs EJ, Hamrick JL (2010) Spatial genetic structure within size classes of the endangered tropical tree Guaiacum sanctum (Zygophyllaceae). Am J Bot 97:1200–1207
García-Verdugo C, Fay MF, Granado-Yela C, Rubio de Casas RR, Balaguer L, Besnard G, Vargas P (2009) Genetic diversity and differentiation processes in the ploidy series of Olea europaea L.: a multiscale approach from subspecies to insular populations. Mol Ecol 18:454–467
García-Verdugo C, Forrest AD, Balaguer L, Fay MF, Vargas P (2010) The relevance of gene flow in metapopulation dynamics of an oceanic island endemic, Olea europaea subsp. guanchica. Evolution 64:3525–3536
Goudet J (2001) FSTAT, version 2.9.3. University of Lausanne. http://www.unil.ch/izea/softwares/fstat.html. Accessed 26 June 2016
Green PS (2002) A revision of Olea L. (Oleaceae). Kew Bull 57:91–140
Guillot G, Mortier F, Estoup A (2005) Geneland: a computer package for landscape genetics. Mol Ecol Notes 5:712–715
Hanski I (1998) Metapopulation dynamics. Nature 396:41–49
Harata T, Nanami S, Yamakura T, Matsuyama S, Chong L, Diway BM, Tan S, Itoh A (2012) Fine-scale spatial genetic structure of ten dipterocarp tree species in a Bornean rain forest. Biotropica 44:586–594
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyze spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Hardy OJ, Maggia L, Bandou E, Breyne P, Caron H, Chevallier M, Doligez A, Dutech C, Kremer A, Latouch-Hallê C, Troispoux V, Veron V, Degen D (2006) Fine-scale genetic structure and gene dispersal inferences in 10 Neotropical tree species. Mol Ecol 15:559–571
Hedrick PW (2005) A standardized genetic differentiation measure. Evolution 59:1633–1638
Henze PB (2000) Layers of time: a history of Ethiopia. Hurst and Company, London
Holderegger R, Buehler D, Gugerli F, Manel S (2010) Landscape genetics of plants. Trends Plant Sci 15:675–683
Jones FA, Hubbell SP (2006) Demographic spatial genetic structure of the Neotropical tree, Jacaranda capaia. Mol Ecol 15:3205–3217
Jordano P, Garcia C, Godoy JA, Garcia-Castano JL (2007) Differential contribution of frugivores to complex seed dispersal patterns. Proc Natl Acad Sci 104:3278–3282
Kaniewski D, Boiy T, Terral JF, Khadari B, Besnard G (2012) Primary domestication and early uses of the emblematic olive tree: palaeobotanical, historical and molecular evidence from the Middle East. Biol Rev 87:885–899
Keller D, Holderegger R, Van Strien MJ, Bolliger J (2015) How to make landscape genetics beneficial for conservation management? Conserv Genet 16:503–512
Latouche-Hallê C, Ramboer A, Bandou E, Caron H, Kremer A (2003) Nuclear and chloroplast genetic structure indicate fine-scale spatial dynamics in a Neotropical tree population. Heredity 91:181–190
Loiselle BE, Sork VL, Nason J, Graham C (1995) Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am J Bot 82:1420–1425
Mills LS, Allendorf FW (1996) The one-migrant-per-generation rule in conservation and management. Conserv Biol 10:1509–1518
Nathan R, Schurr FM, Spiegel O, Steinitz O, Trakhtenbrot A, Tsoar A (2008) Mechanisms of long-distance seed dispersal. Trends Ecol Evol 23:638–647
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nielsen R, Tarpy DR, Reeve HK (2003) Estimating effective paternity number in social insects and the effective number of alleles in a population. Mol Ecol 12:3157–3164
Oddou-Muratorio S, Demesure-Musch B, Pélissier R, Gouyon PH (2004) Impact of gene flow and logging history on the local genetic structure of a scattered tree species, Sorbus torminalis L. Crantz. Mol Ecol 13:3689–3702
Paetkau D, Calvert W, Stirling I, Strobeck C (1995) Microsatellite analysis of population structure in Canadian polar bears. Mol Ecol 4:347–354
Paetkau D, Slade R, Burdens M, Estoup A (2004) Genetic assignment methods for the direct real-time estimation of migration rate: a simulation based exploration of accuracy and power. Mol Ecol 13:55–65
Peakall R, Smouse PE (2012) GenALEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-un updatae. Bioinformatics 28:2537–2539
Petit RJ, El Mousadik A, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conserv Biol 12:844–855
Pinillos V, Cuevas J (2009) Open-pollination provides sufficient level of cross-pollen in Spanish monovarietal olive orchards. Hortscience 44:499–502
Piry S, Alapetite A, Cornuet J-M, Paetkau D, Baudouin L, Estoup A (2004) GENECLASS2: a software for genetic assignment and first-generation migrant detection. J Hered 95:536–539
Raf A, Eva N, Ives VB, Mintesinot B, Martin H, Bart M (2006) Effects of pioneer shrubs on the recruitment of the fleshy-fruited tree Olea europaea ssp. cuspidata in Afromontane savanna. Appl Veg Sci 9:117–126
Rangel TF, Diniz-Filho JAF, Bini LM (2010) SAM: a comprehensive application for spatial analysis in macroecology. Ecography 33:46–50
Sefc KM, Lopes MS, Mendonça D, Dos santos MR, Da Cámara Machado ML, Da Cámara Machado A (2000) Identification microsatellite loci in olive (Olea europaea) and their characterization in Italian and Iberian olive trees. Mol Ecol 9:1171–1193
Sokal RR, Rohlf FJ (1995) Biometry: the principles and practices of statistics in biological research, 3rd edn. WH Freeman and Company, New York
Unger GM, Konrad H, Geburek T (2011) Does spatial genetic structure increase with altitude? An answer from Picea abies in Tyrol, Austria. Plant Syst Evol 292:133–141
Vekemans X, Hardy OJ (2004) New insights from fine-scale spatial genetic structure analyses in plant populations. Mol Ecol 13:921–935
Wang R, Compton SG, Chen XY (2011) Fragmentation can increase spatial genetic structure without decreasing pollen-mediated gene flow in a wind-pollinated tree. Mol Ecol 20:4421–4432
Wassie A, Sterck FJ, Bongers F (2010) Species and structural diversity of church forests in a fragmented Ethiopian highland landscape. J Veg Sci 21:938–948
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Yazdani R, Muona O, Rudin D, Szmidt AE (1985) Genetic structure of a Pinus sylvestris L. seed-tree stand and naturally regenerated understory. Forest Sci 7:430–436
Young A, Boyle T, Brown T (1996) The population genetic consequences of habitat fragmentation for plants. Trends Ecol Evol 11:413–418
Zegeye H, Teketay D, Kelbessa E (2011) Diversity and regeneration status of woody species in Tara Gedam and Abebaye forests, northwestern Ethiopia. J Forest Res 22:315–328
Acknowledgements
We would like to acknowledge the project “Carbon storage and soil biodiversity in forest landscapes in Ethiopia: Knowledge base and participatory management (Carbo part)” for supporting this work. We also thank C. Dobeš and J.P. George for commenting on the manuscript, T. Thalmayr for help with figures, and D. Fentie for assisting in sample collection.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kassa, A., Konrad, H. & Geburek, T. Landscape genetic structure of Olea europaea subsp. cuspidata in Ethiopian highland forest fragments. Conserv Genet 18, 1463–1474 (2017). https://doi.org/10.1007/s10592-017-0993-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-017-0993-z