Abstract
Cryptic invasions are commonly associated with genetic changes of the native species or genetic lineage that the invaders replace. Phenotypic shifts resulting from cryptic invasions are less commonly reported given the relative paucity of historical specimens that document such phenotypic changes. Here, I study such a case in two populations of threespine stickleback from central Europe, comparing contemporary patterns of gene flow with phenotypic changes between historical and contemporary population samples. I find gene flow from an invasive lineage to be associated with significant phenotypic changes, where the degree of phenotypic change corresponds with the level of gene flow that a population receives. These findings underline the utility of combining genetic approaches with phenotypic data to estimate the impact of gene flow in systems where anthropogenic alterations have removed former geographic barriers promoting cryptic invasions.
Similar content being viewed by others
Introduction
Biological invasions commonly pose a threat to native species and even entire ecosystems in their invaded range (Elton 1958; Sakai et al. 2001). The occurrence and impact of such invasions may however be underestimated in cases of cryptic invasions, i.e., when a native species becomes admixed or even genetically replaced by one or multiple distinct genetic lineages of the same species (Saltonstall 2002; Mabuchi et al. 2008; Holsbeek et al. 2008). Cryptic invasions are commonly unrecognized because the invaders are in many cases mistaken for native species (Saltonstall 2002; Mabuchi et al. 2008), yet the invading lineage may differ from their native counterparts in their phenotype (Holsbeek et al. 2008; Mackie et al. 2012) and/or physiology (McIvor et al. 2001). Such phenotypic changes are however less often reported because relatively few cases exist where historical records are available to document phenotypic changes following a cryptic invasion (Wandeler et al. 2007; Crispo et al. 2011).
A great system to assess putative phenotypic changes associated with cryptic invasion is the threespine stickleback (Gasterosteus aculeatus) from central Europe. Freshwater stickleback commonly derive from ancestral marine populations that colonized freshwater habitats since the last glacial period ~12,000 years ago (McKinnon and Rundle 2002). In the River Rhine, stickleback were naturally restricted to as far upstream as Basel and were absent from the Swiss midlands until about 1870 (Fatio 1882; Lucek et al. 2010). Following their subsequent introduction, and facilitated by the channelization of many Swiss waterways, stickleback have since invaded a large part of the country. Genetic analyses of contemporary populations across the invaded range of the country revealed that the colonization of Switzerland was led by three distant genetic lineages from different parts of Europe, which differ especially in their lateral plate phenotypes: The Lake Constance area is dominated by a phenotype that is completely plated along its flank and originates from an East European Baltic lineage (Lucek et al. 2010). In contrast, individuals from the upper Rhone River and the Lake Geneva region have only few lateral plates and derive from a lineage found in the lower Rhone (Lucek et al. 2010). Whereas the aforementioned lineages were introduced in the 1870s, a third and presumably native lineage with few lateral plates, dominates the River Rhine south of Basel (Fatio 1882; Lucek et al. 2010). These distinct lateral plate phenotypes have a simple genetic architecture that is linked to the Ectodysplasin (Eda) locus (Colosimo et al. 2005). Moreover, both the lateral plate phenotypes and the genotypes themselves are known to be under selection (Barrett 2010). For example, piscivorous predators select for an increased lateral plate armor among freshwater fish (Reimchen 1994; Leinonen et al. 2011; Zeller et al. 2012).
Historic records for central Europe (Bertin 1925; Münzing 1963; Gross 1977) and particularly the Basel region (Fatio 1882; Steinmann 1936; Table S1 in supplementary material; see Fig. 1), representing the southernmost part of the native range of stickleback in the River Rhine, suggest that these populations were fixed for a phenotype with only few lateral plates. This is also in agreement with the phylogenetic and biogeographic relationship of these populations (Münzing 1963; Mäkinen and Merilä 2008). In contrast, I observed highly plated phenotypes among contemporary stickleback in this region. Using neutral genetic markers, I test if the presence of such highly plated individuals could reflect cryptic invasion from a distinct lineage into the native range of stickleback, introducing the respective Eda allele. Indeed, the occurrence of highly plated phenotypes within the invaded range of stickleback in Switzerland is linked to gene flow from the genetic lineage that was introduced into the Lake Constance basin (Lucek et al. 2010). However, its potential spread into the native range has so far not been investigated. I expected the potential for cryptic invasion to be high in the Basel region since the geographically close Rhine–Rhone Channel, constructed between 1784 and 1833, connects two main European watersheds: the Mediterranean Sea and the North Sea, each containing distinct stickleback lineages (Mäkinen and Merilä 2008). In addition, the Rhine became connected with the Baltic and the Black Sea drainages further downstream over the last 200 years (Leuven et al. 2009), increasing the potential for intraspecific gene flow from highly plated East European stickleback populations (Münzing 1963). By including many populations from across Europe, I aim to identify the potential sources for gene flow. Lastly, I compare the phenotypes of historical populations from the Basel region that predate the introduction and expansion of nonnative stickleback lineages in Switzerland with contemporary samples collected at the same sites. With this comparison, I am able to infer the effects of gene flow on the phenotypic composition of the two populations for which historical data were available. I predict the degree of phenotypic changes through time to reflect the relative level of gene flow and admixture that a population receives.
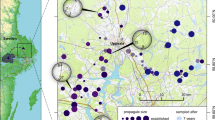
Overview of the studied populations and their associated genetic structure: a Map of Europe depicting the location of the 18 populations included in this study. The inset focuses on the region around Basel. b Genetic structure among populations based on individual-based Bayesian assignments using the program Structure. Shown is the best statistical clustering, assuming seven genetic clusters. Numbers indicate the population ID (see Table 1 for details). Pie plots indicate the allele frequency of the two Stn382 alleles linked to the Eda gene (white—L allele, black—C allele)
Materials and methods
Two historical stickleback populations from the Basel region dating back to the 1870s were available from the natural history museums in Basel and Vienna (NBasel = 11—sampled in 1875, NVillage-Neuf = 28—sampled in 1879). I collected stickleback at the same locations as described in the museum records (NBasel = 27—sampled in 2010, NVillage-Neuf = 26—sampled in 2012). Hereafter I refer to these two types of samples as “historical” or “contemporary” respectively. The samples from the Basel museum were collected in the River Wiese in Basel (Fig. 1a), a tributary to the Rhine. Samples from Village-Neuf stem from a man-made canal built in the 1830s, which connects the Rhine–Rhone Channel with the Rhine close to Basel. The canal of Village-Neuf and the Wiese are separated by a ~250 m stretch of the Rhine. The Village-Neuf site contains several water locks and water velocity is lower than in the lower stretch of the Wiese, which contains consecutive artificial ramps (Lucek personal observation). To infer potential contemporary gene flow and to determine its possible origin, I included stickleback from 16 additional European populations that cover the putative ancestral lineages of invasive stickleback in Switzerland (Lucek et al. 2010), parts of the native range of the upper Rhine and the potential lineages to which native stickleback in the River Rhine have become connected (Leuven et al. 2009). All contemporary individuals were collected using hand nets and minnow traps and were preserved in 70 % ethanol. A fin clip was separately taken and stored in absolute ethanol for the genetic analyses.
Due to the type of preservation and access restrictions, none of the historical specimens were available for DNA extraction or invasive handling that would have, for example, allowed me to determine the sex of the fish. Consequently, I focused on changes in the lateral plate phenotype as these were not sexually dimorphic among contemporary individuals (ANOVA: Basel—F 1,25 = 0.01, p = 0.991; Village-Neuf—F 1,24 = 0.38, p = 0.544). I counted all lateral plates, including the keel plates, on the left flank for both historical and contemporary specimens from Basel and Village-Neuf. Additional lateral plate phenotypes from the same time period were available for 40 individuals from the Basel site (Fatio 1882; Table S1 in supplementary material). I compared the number of lateral plates among contemporary and historic (including additional phenotypes from Fatio) samples using Kruskal–Wallis rank sum tests because plate numbers were not normally distributed (Fig. 2). Moreover, I compared the proportion of individuals with more than 10 lateral plates (hereafter referred to as being “highly plated”) between historical and contemporary samples from the same site using a two-sample test for equality of proportions. I used R 3.1.2 (R Development Core Team 2014) to conduct all statistical analyses.
In total, I genotyped 388 individuals from 18 contemporary stickleback populations at nine microsatellite loci. These loci have been shown to be neutral in several European populations (Lucek et al. 2014a, b; Lucek and Seehausen 2015). The data for eight populations overlaps with previous studies (Table 1). I extracted DNA for all individuals using a 10 % Chelex solution following the manufacturer’s protocol (Biorad, California, USA). Detailed information on the marker identity, the multiplexing setup and the PCR protocol are given in Lucek et al. (2014b). Alleles were visualized on an ABI 3130XL and scored with GeneMapper 4.0 (Applied Biosystems, Switzerland). I subsequently assessed the potential for gene flow among all contemporary populations using an admixture model implemented in Structure 2.3.3 (Falush et al. 2007) with 10,000 burn-in steps followed by 100,000 MCMC steps. The simulation was performed assuming 1–10 genetic clusters (K) with 10 replicates for each K. I then determined the optimal number of clusters based on the log likelihood of each run and its variation among runs for the same K (Evanno et al. 2005). To estimate the genetic variation within a population, I calculated the observed (H O ) and expected (H E ) heterozygosity as well as the effective number of alleles (A R ), i.e. the number of alleles weighted for their frequency in Genodive 2.0 (Meirmans and Van Tienderen 2004). I further genotyped 426 contemporary individuals for Stn382 (Table 1), a marker flanking a 60 bp indel in intron 1 of the Eda gene. This marker yields either a 158 bp (‘‘Eda L’’) allele, associated with a phenotype that has less than ten lateral plates on either side or a 218 bp (‘‘Eda C’’) allele. The latter is associated with a completely plated phenotype, where heterozygous individuals have generally more than ten lateral plates on either side (Colosimo et al. 2005). PCR products were analyzed on a 1.5 % agarose gel, stained with ethidium bromide, where bands were scored by eye. To test if the number of lateral plates is associated with the number of Eda C alleles, I employed a linear model following Colosimo et al. (2005). Lastly, I tested for deviations from a Hardy–Weinberg equilibrium (HWE) in Genodive using 10,000 bootstrap replicates separately for the microsatellite markers and the Eda genotype.
Results and discussion
Cryptic invasions are difficult to detect, consequently their impact on native populations is often underestimated (Saltonstall 2002). The potential for phenotypic shifts in native populations as a result of a cryptic invasion is rarely assessed (Wandeler et al. 2007; Crispo et al. 2011; but see Holsbeek et al. 2008; Mackie et al. 2012), which may also reflect the paucity of historic samples and data available to document such shifts. Comparing historical and contemporary stickleback, I show a phenotypic shift in lateral plate characteristics, a trait which is associated with a major effect gene (Fig. 2): The proportion of highly plated individuals increased significantly between the historical and contemporary samples (Basel: \(\chi_{1}^{2} = { 7}. 2 4\), p = 0.007; Village-Neuf: \(\chi_{1}^{2} = { 23}. 7 7\), p < 0.001), which was also true for the average number of lateral plates in Village-Neuf (\(\chi_{1}^{2} = { 17}. 8 6\), p < 0.001), but not Basel (\(\chi_{1}^{2} = { 3} . 0 2\), p = 0.082). Contemporary Village-Neuf specimens have a higher plate number than their contemporary Basel counterparts (\(\chi_{1}^{2} = { 9} . 7 7\), p = 0.002), while the two populations did not differ historically (\(\chi_{1}^{2} = \, 0.31\), p = 0.575). The occurrence of highly plated stickleback is significantly correlated with the presence of the Eda C allele among contemporary individuals (Basel: R 2 = 0.794, F 1,25 = 96.19, p < 0.001; Village-Neuf: R 2 = 0.818, F 1,24 = 108.10, p < 0.001) and Eda explains a comparable level of the phenotypic variation as for stickleback in other parts of their native range (Colosimo et al. 2005).
The presence of highly plated individuals within the invasive range of stickleback in Switzerland are a result of gene flow with the genetic lineage that was introduced into Lake Constance (Lucek et al. 2010). Concomitantly, the Structure analyses conducted in this study identified the populations in the Lake Constance region as the most likely source for gene flow into the native range of stickleback in the Rhine north of Basel, with a smaller contribution from the upper Rhone region (Fig. 1b). Gene flow was highest at the sites Village-Neuf and the connected Rhine–Rhone Channel, where several individuals were almost completely assigned to the genetic cluster found in the Lake Constance region (Fig. 1b). This potentially reflects migration or the deliberate introduction of Lake Constance derived stickleback in the native region around Basel. The subsequent cryptic invasion likely increased heterozygosity and hence the level of standing genetic variation (Table 1). The other genetic clusters identified by Structure reflect distinct geographic regions within watersheds (Fig. 1b). Specifically, individuals from the Basel region, the upper Doubs/Rhone drainage and the Basel site form a distinct genetic cluster, likely representing the ancestral local gene pool for this region.
The degree of phenotypic differentiation between historical and contemporary samples (Fig. 2) corresponds to the relative degree of gene flow observed between the Lake Constance lineage and contemporary specimens from the Basel region (Fig. 1). This is consistent with theoretical predictions (Räsänen and Hendry 2008) and suggests that increased gene flow and admixture resulted in phenotypic shifts, which may even override local selection on lateral plate phenotypes (Raeymaekers et al. 2014; Ferchaud and Hansen 2016). Gene flow and subsequent admixture, rather than the coexistence of distinct lineages, is moreover indicated by the absence of deviations from Hardy–Weinberg equilibrium for both Eda and the microsatellite markers (Waples 2015). Only two out of 162 comparisons deviated significantly from equilibrium following a sequential Bonferroni correction (Stn130: L’Isle-sur-la-Sorgue; Stn173: Tübingen). However, despite the low level of introgression at neutral markers, highly plated individuals and the associated Eda C allele occur at the Basel site in the present day. Selection against immigrants and/or hybrids, as well as the presence of physical barriers such as the ramps in the River Wiese, could reduce the potential for gene flow (Weibel and Peter 2012). This could have resulted in the persistence of the putatively ancestral gene pool at neutral markers in the Basel site. Introgression at a phenotype-linked marker may nevertheless occur if the phenotype experiences positive selection (Rieseberg 2011). This has been suggested to be the case for Eda in some stickleback populations within their invaded range in Switzerland (Lucek et al. 2014a). Indeed, lateral plate number and Eda itself can be under selection (Barrett 2010; Leinonen et al. 2011; Zeller et al. 2012). An increased number of lateral plates and Eda C alleles could reflect a change in the local predator community towards an increased predation regime (Reimchen 1994). However, no historical data is available for the predator species composition, limiting speculation about environmental changes over this time period. Similarly, the current data does not allow me to infer if the observed phenotypic changes are maladaptive or adaptive, which would require further experimental testing (Kawecki and Ebert 2004).
Taken together, my findings stress the importance of combining genetic and phenotypic approaches to document the occurrence and extent of cryptic invasions (Bacigalupe 2009). Phenotypic shifts as a consequence of introgression at phenotype-linked markers are likely to be underestimated by neutral markers alone. This was highlighted in this study by contrasting changes in the Eda allele frequencies and neutral markers (Fig. 1). However, cryptic invasions resulting in phenotypic shifts could be common given the vast scale at which natural boundaries to gene flow are removed by humans, connecting formerly isolated biogeographic regions (Leuven et al. 2009).
References
Bacigalupe LD (2009) Biological invasions and phenotypic evolution: a quantitative genetic perspective. Biol Invasions 11:2243–2250. doi:10.1007/s10530-008-9411-2
Barrett RDH (2010) Adaptive evolution of lateral plates in three-spined stickleback Gasterosteus aculeatus: a case study in functional analysis of natural variation. J Fish Biol 77:311–328. doi:10.1111/j.1095-8649.2010.02640.x
Bertin L (1925) Recherches bionomiques, biométriques et sysématiques sur les épinoches (Gastérostéidés). Ann Inst Océanogr Monaco 2:1–204
Colosimo PF, Hosemann KE, Balabhadra S et al (2005) Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science 307:1928–1933. doi:10.1126/science.1107239
Crispo E, Moore J-S, Lee-Yaw JA et al (2011) Broken barriers: human-induced changes to gene flow and introgression in animals: an examination of the ways in which humans increase genetic exchange among populations and species and the consequences for biodiversity. BioEssays 33:508–518. doi:10.1002/bies.201000154
Elton CS (1958) The ecology of invasions by animals and plants. University Of Chicago Press, Chicago
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. doi:10.1111/j.1365-294X.2005.02553.x
Falush D, Stephens M, Pritchard J (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578. doi:10.1111/j.1471-8286.2007.01758.x
Fatio V (1882) Faune des vertébrés de la Suisse. H. Georg, Genève
Ferchaud A-L, Hansen MM (2016) The impact of selection, gene flow and demographic history on heterogeneous genomic divergence: three-spine sticklebacks in divergent environments. Mol Ecol 25:238–259. doi:10.1111/mec.13399
Gross HP (1977) Adaptive trends of environmentally sensitive traits in the three-spined stickleback, Gasterosteus aculeatus L. J Zool Syst Evol Res 15:252–278
Holsbeek G, Mergeay J, Hotz H et al (2008) A cryptic invasion within an invasion and widespread introgression in the European water frog complex: consequences of uncontrolled commercial trade and weak international legislation. Mol Ecol 17:5023–5035. doi:10.1111/j.1365-294X.2008.03984.x
Kawecki T, Ebert D (2004) Conceptual issues in local adaptation. Ecol Lett 7:1225–1241. doi:10.1111/j.1461-0248.2004.00684.x
Leinonen T, Herczeg G, Cano JM, Merilä J (2011) Predation-imposed selection on threespine stickleback (Gasterosteus aculeatus) morphology: a test of the refuge use hypothesis. Evolution 65:2916–2926. doi:10.1111/j.1558-5646.2011.01349.x
Leuven RSEW, van der Velde G, Baijens I et al (2009) The river Rhine: a global highway for dispersal of aquatic invasive species. Biol Invasions 11:1989–2008. doi:10.1007/s10530-009-9491-7
Lucek K, Seehausen O (2015) Distinctive insular forms of threespine stickleback (Gasterosteus aculeatus) from western Mediterranean islands. Conserv Genet 16:1319–1333. doi:10.1007/s10592-015-0742-0
Lucek K, Roy D, Bezault E et al (2010) Hybridization between distant lineages increases adaptive variation during a biological invasion: stickleback in Switzerland. Mol Ecol 19:3995–4011. doi:10.1111/j.1365-294X.2010.04781.x
Lucek K, Lemoine M, Seehausen O (2014a) Contemporary ecotypic divergence during a recent range expansion was facilitated by adaptive introgression. J Evol Biol 27:2233–2248. doi:10.1111/jeb.12475
Lucek K, Sivasundar A, Kristjánsson BK et al (2014b) Quick divergence but slow convergence during ecotype formation in lake and stream stickleback pairs of variable age. J Evol Biol 27:1878–1892. doi:10.1111/jeb.12439
Mabuchi K, Senou H, Nishida M (2008) Mitochondrial DNA analysis reveals cryptic large-scale invasion of non-native genotypes of common carp (Cyprinus carpio) in Japan. Mol Ecol 17:796–809. doi:10.1111/j.1365-294X.2007.03626.x
Mackie JA, Darling JA, Geller JB (2012) Ecology of cryptic invasions: latitudinal segregation among Watersipora (Bryozoa) species. Sci Rep 2:1–10. doi:10.1038/srep00871
Mäkinen HS, Merilä J (2008) Mitochondrial DNA phylogeography of the three-spined stickleback (Gasterosteus aculeatus) in Europe—evidence for multiple glacial refugia. Mol Phylogenet Evol 46:167–182. doi:10.1016/j.ympev.2007.06.011
McIvor L, Maggs CA, Provan J, Stanhope MJ (2001) rbcL sequences reveal multiple cryptic introductions of the Japanese red alga Polysiphonia harveyi. Mol Ecol 10:911–919
McKinnon JS, Rundle H (2002) Speciation in nature: the threespine stickleback model systems. Trends Ecol Evol 17:480–488
Meirmans PG, Van Tienderen P (2004) GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes 4:792–794
Münzing J (1963) The evolution of variation and distributional patterns in European populations of the three-spined stickleback, Gasterosteus aculeatus. Evolution 17:320–332
R Development Core Team (2014) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.r-project.org
Raeymaekers JAM, Konijnendijk N, Larmuseau MHD et al (2014) A gene with major phenotypic effects as a target for selection vs. homogenizing gene flow. Mol Ecol 23:162–181. doi:10.1111/mec.12582
Räsänen K, Hendry AP (2008) Disentangling interactions between adaptive divergence and gene flow when ecology drives diversification. Ecol Lett 11:624–636. doi:10.1111/j.1461-0248.2008.01176.x
Reimchen TE (1994) Predators and morphological evolution in threespine stickleback. Predators and morphological evolution in threespine stickleback. In: Bell M, Foster S (eds) The evolutionary biology of the threespine stickleback, 1st edn. Oxford University Press, Oxford, pp 240–276
Rieseberg L (2011) Adaptive introgression: the seeds of resistance. Curr Biol 21:R581–R583. doi:10.1016/j.cub.2011.06.038
Sakai A, Allendorf F, Holt J et al (2001) The population biology of invasive species. Annu Rev Ecol Evol Syst 32:305–332
Saltonstall K (2002) Cryptic invasion by a non-native genotype of the common reed, Phragmites australis, into North America. Proc Natl Acad Sci USA 99:2445–2449. doi:10.1073/pnas.032477999
Steinmann P (1936) Die Fische der Schweiz. Sauerländer, Aarau
Wandeler P, Hoeck PEA, Keller LF (2007) Back to the future: museum specimens in population genetics. Trends Ecol Evol 22:634–642. doi:10.1016/j.tree.2007.08.017
Waples RS (2015) Testing for Hardy–Weinberg proportions: have we lost the plot? J Hered 106:1–19. doi:10.1093/jhered/esu062
Weibel D, Peter A (2012) Effectiveness of different types of block ramps for fish upstream movement. Aquat Sci 75:251–260. doi:10.1007/s00027-012-0270-7
Zeller M, Lucek K, Haesler MP et al (2012) Signals of predation-induced directional and disruptive selection in the threespine stickleback. Evol Ecol Res 14:193–205
Acknowledgments
I would like to thank Mélissa Lemoine, Timothy J. Alexander, Rebecca Best, Juha Merilä and David A. Marques for valuable comments on earlier versions of the manuscript. I am further indebted to Ole Seehausen, Guy Périat, Rafał Bernaś and David A. Marques for providing specimens and Helmut Wellendorf from the Museum of Natural History in Vienna as well as Urs Wüest from the Natural History Museum of Basel for their assistance in obtaining historical phenotypic data. I dedicate this paper to my mother Claudia Daniela Lucek-Schneider (1961–2009), who financially supported my data collection at the Museum of Natural History in Vienna. This research was funded by a Swiss National Science Foundation Early Postdoc.Mobility grant P2BEP3_152103.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Lucek, K. Cryptic invasion drives phenotypic changes in central European threespine stickleback. Conserv Genet 17, 993–999 (2016). https://doi.org/10.1007/s10592-016-0837-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-016-0837-2