Abstract
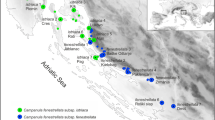
Lepidium papilliferum of southwest Idaho was previously treated as an infraspecific variety of Lepidium montanum. Chloroplast (cpDNA) sequences, nuclear ribosomal internal transcribed spacer (ITS) sequences, and AFLPs were used to test species delimitations and other possible evolutionarily significant units (ESU) based on genetic differentiation, isolation by distance (IBD), and genetic admixture among 32 L. montanum and 21 L. papilliferum collections from the western US. The L. papilliferum AFLP genotypes formed a monophyletic clade. However, the AFLP genotypes of L. montanum samples from eight western sites were more similar to L. papilliferum, which together comprise a regionally significant West clade showing significant differentiation from eastern L. montanum collections (East clade). Bayesian analysis of AFLP genotypes detected possible admixture between L. papilliferum and related western L. montanum collections. Neither taxa nor regionally significant AFLP clades displayed reciprocally monophyletic cpDNA or ITS sequences, but the AFLP clades showed stronger cpDNA differentiation and unique ITS alleles. The East and West clades fit models of speciation with relatively strong IBD within groups and weak IBD between groups, based on correlations between the average number of AFLP differences and geographic distances among collection sites, but comparisons between taxa did not fit this model. Conversely, relatively strong partial correlations between AFLP and taxonomic differences, controlling for geography, support taxonomic delimitations. Results suggest that L. papilliferum is a distinct subgroup of L. montanum influenced by speciation. However, gene flow or common ancestry between L. papilliferum and western forms of L. montanum provide a basis for other possible ESUs.







Similar content being viewed by others
References
Álvarez I, Wendel JF (2003) Ribosomal ITS sequences and plant phylogenetic inference. Mol Phylogenet Evol 29:417–434
Avise JC, Neigle JE, Arnold J (1984) Demographic influences on mitochondrial DNA lineage survivorship in animal populations. J Mol Evol 20:99–105
Baldwin BG (1992) Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants: an example from the Compositae. Mol Phylogenet Evol 1:3–16
Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ (1995) The ITs region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Ann Missouri Bot Gard 82:247–277
Bohonak AJ, Davies N, Roderick GK, Villablanca F (1998) Is population genetics mired in the past? Trends Ecol Evol 13:360
Bossert JL, Prowell DP (1998) Genetic estimates of population structure and gene flow: limitations, lessons and new directions. TREE 13:202–205
Bowman JL, Brüggemann H, Lee JY, Mummenhoff K (1999) Evolutionary changes in floral structure within Lepidium L. (Brassicaceae). Int J Plant Sci 160:917–929
Bulgin NL, Gibbs HL, Vickery P, Baker AJ (2003) Ancestral polymorphisms in genetic markers obscure detection of evolutionarily distinct populations in the endangered Florida grasshopper sparrow (Ammodramus savannarum floridanus). Mol Ecol 12:8331–8844
Clark AG (1990) Inference of haplotypes from PCR-amplified samples of diploid populations. Mol Biol Evol 72:111–122
Culumber CM (2007) DNA barcoding of western North American taxa Leymus (Poaceae) and Lepidium (Brassicaceae). MS thesis, Utah State University
Davis RJ (1952) Flora of Idaho. Brigham Young University Press, Provo, Utah, p 347
Despres L, Gielly L, Redoutet W, Taberlet P (2003) Using AFLP to resolve phylogenetic relationships in a morphologically diversified plant species complex when nuclear and chloroplast sequences fail to reveal variability. Mol Phylogenet Evol 27:185–196
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
Good DA, Wake DB (1992) Geographic variation and speciation in the torrent salamanders of the genus Rhyacotriton (Caudata: Rhyacotritonidae). Univ Calif Publ Zool 126:1–91
Hitchcock CL (1950) On the subspecies of Lepidium montanum. Madroño 10:155–158
Holmgren NH, Holmgren PK, Cronquist A (2005) Intermountain flora, vascular plants of the intermountain west, USA, vol 2, Part B, Subclass Dilleniideae. The New York Botanical Garden Press, New York
Hutchison DW, Templeton AR (1999) Correlation of pairwise genetic and geographic distance measures: inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution 53:1898–1914
Jakob SS, Blattner FR (2006) A chloroplast genealogy of Hordeum (Poaceae): long-term persisting haplotypes, incomplete lineage sorting, regional extinction, and the consequences for phylogenetic inference. Mol Biol Evol 23:1602–1612
Jensen JL, Bohonak AJ, Kelley ST (2005) Isolation by distance, web service. BMC Genet 6:13
Jones TA, Larson SR, Wilson BL (2008) Genetic differentiation and admixture among Festuca idahoensis, F. roemeri, and F. ovina detected in AFLP, ITS, and cpDNA. Bot 86:422–434
Jump AS, Woodward FI, Burke T (2003) Cirsium species show disparity in patterns of genetic variation at their range-edge, despite similar patterns of reproduction and isolation. New Phytol 160:359–370
Kimura M (1981) Estimation of evolutionary distances between homologous nucleotide sequences. Proc Natl Acad Sci USA 78:454–458
Koopman WJM (2005) Phylogenetic signal in AFLP data sets. Syst Biol 54:197–217
Koopman WJM, Wisseman V, Cock KD, Huylenbroeck JV, Riek JD, Sabatino GJH, Visser D, Vosman B, Ritz CM, Maes B, Werlemark G, Nybom H, Debener T, Linde M, Smulders MJM (2008) AFLP markers as a tool to reconstruct complex relationships: a case study in Rosa (Rosaceae). Am J Bot 95:353–366
Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH (2005) Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci USA 102:8369–8374
Kropf M, Kadereit JW, Comes HP (2002) Late Quaternary distributional stasis in the submediterranean mountain plant Anthyllis montana L. (Fabaceae) inferred from ITS sequences and amplified fragment length polymorphism markers. Mol Ecol 11:447–463
Lee JY, Mummenhoff K, Bowman JL (2002) Allopolyploidization and evolution of species with reduced floral structures in Lepidium L. (Brassicaceae). Proc Natl Acad Sci USA 99:16835–16840
Leonard AC, Franson SD, Hertzberg VS, Smith MK, Toth GP (1999) Hypothesis testing with the similarity index. Mol Ecol 8:2105–2114
Lexer C, Kremer A, Petit RJ (2006) Shared alleles in sympatric oaks: recurrent gene flow is a more parsimonious explanation than ancestral polymorphism. Mol Ecol 15:2007–2012
Manel S, Gaggiotti OE, Waples RS (2005) Assignment methods: matching biological questions with appropriate techniques. Trends Ecol Evol 20:136–142
Meyer SD, Allen PS (2005) Lepidium papilliferum soil and seed bank characterization on the orchard training area. US Forest Service, Intermountain Research Station, Shrub Sciences Laboratory, Brigham Young University, Provo, Utah
Meyer SD, Quinney D, Weaver J (2006) A stochastic population model for Lepidium papilliferum (Brassicaceae), a rare desert ephemeral with a persistent seed bank. Am J Bot 93:891–902
Modliszewski JL, Thomas DT, Fan C, Crawford DJ, DePamphilis CW, Xiang Q-YJ (2006) Ancestral chloroplast polymorphism and historical secondary contact in a broad hybrid zone of Aesculus (Sapindaceae). Am J Bot 93:377–388
Moritz C (1994) Defining ‘evolutionarily significant units’ for conservation. TREE 9:373–375
Moyle LC (2006) Correlates of genetic differentiation and isolation by distance in 17 congeneric Silene species. Mol Ecol 15:1067–1081
Muir G, Schlötterer C (2005) Evidence for shared ancestral polymorphisms rather than recurrent gene flow at microsatellite loci differentiating two hybridizing oaks (Quercus spp.). Mol Ecol 14:549–561
Mummenhoff K, Linder P, Friesen N, Bowman JL, Lee JY, Franzke A (2004) Molecular evidence for biocontinentaly hybridogenous genomic constitution in Lepidium sensu stricto (Brassicaceae) species from Australia and New Zealand. Am J Bot 91:254–261
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Neigel JE, Avise JC (1986) Phylogenetic relationships of mitochondrial DNA under various demographic models of speciation. In: Nevo E, Karlin S (eds) Evolutionary processes and theory. Academic Press, New York, pp 515–534
Ouborg NJ, Piquot Y, Van Groenendael JM (1999) Population genetics, molecular markers and the study of dispersal in plants. J Ecol 87:551–568
Petit R, Duminil J, Fineschi S et al (2005) Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol Ecol 14:689–701
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Puorto G, Salomão M, Graca Da, Theakston RDG, Thorpe RS, Warrell DA, Wűster W (2001) Combining mitochondrial DNA sequences and morphological data to infer species boundaries: phylogeography of lanceheaded pitvipers in the Brazilian Atlantic forest, and the status of Bothrops pradoi (Squamata: Serpentes: Viperideae). J Evol Biol 14:527–538
Rollins RC (1993) The Cruciferae of continental North America. Stanford University Press, USA
Rollins RC, Rudenberg L (1977) Chromosome numbers of Cruciferae III. Contrib Gray Herb Harvard Univ 207:101–116
Scotti I, Vendramin GG, Matteotti LS, Sarponi C, Sari-Gorla M, Binelli G (2000) Postglacial recononization routes for Piceae abies K. in Italy as suggested by the analysis of sequence-characterized amplified region (SCAR) markers. Mol Ecol 9:699–708
Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, Miller J, Siripun KC, Winder CT, Schilling EE, Small RL (2005) The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am J Bot 92:142–166
Sites JW, Marshall JC (2003) Delimiting species: a Renaissance issue in systematic biology. Trends Ecol Evol 18:462–469
Sites JW, Marshall JC (2004) Operational criteria for delimiting species. Annu Rev Ecol Evol Syst 35:199–227
Slatkin M (1985) Rare alleles as indicators of gene flow. Evolution 39:53–65
Slatkin M (1993) Isolation by distance in equilibrium and non-equilibrium populations. Evolution 47:264–279
Stillman AJ (2006) Population genetics and mating system of the rare polyploid Lepidium papilliferum (Brassicaceae), a southwestern Idaho endemic. MS thesis, Boise State University
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of cpDNA. Plant Mol Biol 17:1105–1109
US Fish and Wild life Service (2006) Draft best available biological information for slickspot peppergrass (Lepidium papilliferum). Snake River Fish and Wildlife Office, Boise, Idaho
US Fish and Wild life Service (2007) Endangered and threatened wildlife and plants; withdrawl of proposed rule to list Lepidium palliferum (slickspot peppergrass). Fed Reg 72:1621–1644
US Fish and Wildlife Service (1990) Endangered and threatened wildlife and plants; review of plant taxa for listing as endangered or threatened species. Fed Reg 55:6181–6229
US Fish and Wildlife Service (2002) Endangered and threatened wildlife and plants; listing the plant Lepidium papilliferum (slickspot peppergrass) as endangered. Fed Reg 67:46441–46450
Vos P, Hogers R, Bleeker M, Reijans M, Van De Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Waples RS (1991) Pacific salmon, Oncorhynchus spp., and the definition of ‘species’ under the endangered species act. Mar Fish Rev 53:11–22
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, California, pp 315–322
Whitlock MC, McCauley DE (1999) Indirect measures of gene flow and migration: FST ≠ 1/(4Nm + 1). Heredity 82:117–125
Wolfe KH, Li W-H, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA 84:9054–9058
Wright S (1931) Evolution in Mendelian populations. Genetics 16:97–159
Wright S (1943) Isolation by distance. Genetics 28:139–156
Wright S (1978) Evolution and the genetics of populations, Vol. 4. Variability within and among natural populations. University of Chicago Press, Chicago, IL, USA
Wu X-L, Larson SR, Hu Z-M, Palazzo AJ, Jones TA, Wang RR-C, Jensen KB, Chatterton NJ (2003) Molecular genetic linkage maps for allotetraploid Leymus wildryes (Gramineae: Triticeae). Genome 46:627–646
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Larson, S.R., Culumber, C.M., Schweigert, R.N. et al. Species delimitation tests of endemic Lepidium papilliferum and identification of other possible evolutionarily significant units in the Lepidium montanum complex (Brassicaceae) of western North America. Conserv Genet 11, 57–76 (2010). https://doi.org/10.1007/s10592-009-0002-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-009-0002-2