Abstract
Microgametogenesis in angiosperms results in two structurally and functionally different cells, one generative cell, which subsequently forms the sperm cells, and the vegetative cell. We analysed the chromatin properties of both types of nuclei after first and second pollen mitosis in rye (Secale cereale). The condensed chromatin of generative nuclei is earmarked by an enhanced level of histone H3K4/K9 dimethylation and H3K9 acetylation. The less condensed vegetative nuclei are RNA polymerase II positive. Trimethylation of H3K27 is not involved in transcriptional downregulation of genes located in generative nuclei as H3K27me3 was exclusively detected in the vegetative nuclei. The global level of DNA methylation does not differ between both types of pollen nuclei. In rye, unlike in Arabidopsis thaliana (Ingouff et al. Curr Biol 17:1032–1037 2007; Schoft et al. EMBO Rep 10:1015–1021 2009), centromeric histone H3 is not excluded from the chromatin of the vegetative nucleus and the condensation degree of centromeric and subtelomeric regions did not differ between the generative and vegetative nuclei. Differences between rye and A. thaliana data suggest that the chromatin organization in mature nuclei of pollen grains is not universal across angiosperms.




Similar content being viewed by others
Abbreviations
- CENH3:
-
Centromeric histone H3
- ChIP:
-
Chromatin immunoprecipitation
- DAPI:
-
4′,6-diamidino-2-phenylindole
- FISH:
-
Fluorescence in situ hybridization
- H3K4me2:
-
Dimethylated histone H3 at lysine 4
- H3K9me2:
-
Dimethylated histone H3 at lysine 9
- H3K27me3:
-
Trimethylated histone H3 at lysine 27
- H3K9ac:
-
Acetylated histone H3 at lysine 9
- Sup:
-
Supernatant
- Pel:
-
Pellet
References
Borg M, Brownfield L, Twell D (2009) Male gametophyte development: a molecular perspective. J Exp Bot 60:1465–1478
Carchilan M, Delgado M, Ribeiro T, Costa-Nunes P, Caperta A, Morais-Cecilio L, Jones RN, Viegas W, Houben A (2007) Transcriptionally active heterochromatin in rye B chromosomes. Plant Cell 19:1738–1749
Cartagena JA, Matsunaga S, Seki M, Kurihara D, Yokoyama M, Shinozaki K, Fujimoto S, Azumi Y, Uchiyama S, Fukui K (2008) The Arabidopsis SDG4 contributes to the regulation of pollen tube growth by methylation of histone H3 lysines 4 and 36 in mature pollen. Dev Biol 315:355–368
Dunwell JM (2010) Haploids in flowering plants: origins and exploitation. Plant Biotechnol J 8:377–424
Francki MG (2001) Identification of Bilby, a diverged centromeric Ty1-copia retrotransposon family from cereal rye (Secale cereale L.). Genome 44:266–274
Fuchs J, Demidov D, Houben A, Schubert I (2006) Chromosomal histone modification patterns—from conservation to diversity. Trends Plant Sci 11:199–208
Gerlach WL, Bedbrook JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res 7:1869–1885
Honys D, Twell D (2003) Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol 132:640–652
Houben A, Demidov D, Gernand D, Meister A, Leach CR, Schubert I (2003) Methylation of histone H3 in euchromatin of plant chromosomes depends on basic nuclear DNA content. Plant J 33:967–973
Houben A, Orford SJ, Timmis JN (2006) In situ hybridization to plant tissues and chromosomes. Meth Mol Biol 326:203–218
Houben A, Demidov D, Caperta AD, Karimi R, Agueci F, Vlasenko L (2007) Phosphorylation of histone H3 in plants–a dynamic affair. Biochim Biophys Acta 1769:308–315
Ingouff M, Hamamura Y, Gourgues M, Higashiyama T, Berger F (2007) Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr Biol 17:1032–1037
Ingouff M, Rademacher S, Holec S, Soljic L, Xin N, Readshaw A, Foo SH, Lahouze B, Sprunck S, Berger F (2010) Zygotic resetting of the histone 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr Biol 20:2137–2143
Janousek B, Zluvova J, Vyskot B (2000) Histone H4 acetylation and DNA methylation dynamics during pollen development. Protoplasma 211:116–122
Jimenez MM, Romera F, Puertas MJ, Jones RN (1994) B-Chromosomes in inbred lines of rye (Secale cereale L).1. Vigor and fertility. Genetica 92:149–154
Kim M, Kim J, Yoon M, Choi DI, Lee KM (2004) Origin of multicellular pollen and pollen embryos in cultured anthers of pepper (Capsicum annuum). Plant Cell Tissue Organ Cult 77:63–72
Kornberg RD (1999) Eukaryotic transcriptional control. Trends Biochem Sci 24:M46–M49
Ma R, Guo Y, Pulli S (2004) Comparison of anther and microspore culture in the embryogenesis and regeneration of rye (Secale cereale). Plant Cell Tissue Organ Cult 76:147–157
Maraschin SF, de Priester W, Spaink HP, Wang M (2005) Androgenic switch: an example of plant embryogenesis from the male gametophyte perspective. J Exp Bot 56:1711–1726
Mehta GD, Agarwal MP, Ghosh SK (2010) Centromere identity: a challenge to be faced. Mol Genet Genomics 284:75–94
Nagaki K, Murata M (2005) Characterization of CENH3 and centromere-associated DNA sequences in sugarcane. Chromosome Res 13:195–203
Nagaki K, Talbert PB, Zhong CX, Dawe RK, Henikoff S, Jiang JM (2003) Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics 163:1221–1225
Nagaki K, Cheng ZK, Ouyang S, Talbert PB, Kim M, Jones KM, Henikoff S, Buell CR, Jiang JM (2004) Sequencing of a rice centromere uncovers active genes. Nat Genet 36:138–145
Oakeley EJ, Podesta A, Jost JP (1997) Developmental changes in DNA methylation of the two tobacco pollen nuclei during maturation. Proc Natl Acad Sci USA 94:11721–11725
Okada T, Singh MB, Bhalla PL (2006) Histone H3 variants in male gametic cells of lily and H3 methylation in mature pollen. Plant Mol Biol 62:503–512
Okada T, Singh MB, Bhalla PL (2007) Transcriptome profiling of Lilium longiflorum generative cells by cDNA microarray. Plant Cell Rep 26:1045–1052
Ravi M, Chan SW (2010) The rapidely evolving centromere-specific histone has stringent functional requirements in Arabidopsis thaliana. Genetics 186:461–471
Reynolds TL (1993) A cytological analysis of microspores of Triticum aestivum (Poaceae) during normal ontogeny and induced embryogenic development. Am J Bot 80:569–576
Ribeiro T, Viegas W, Morais-Cecilio L (2009) Epigenetic marks in the mature pollen of Quercus suber L. (Fagaceae). Sex Plant Reprod 22:1–7
Roudier F, Teixeira FK, Colot V (2009) Chromatin indexing in Arabidopsis: an epigenomic tale of tails and more. Trends Genet 25:511–517
Sano Y, Tanaka I (2010) Distinct localization of histone H3 methylation in the vegetative nucleus of lily pollen. Cell Biol Int 34:253–259
Schoft VK, Chumak N, Mosiolek M, Slusarz L, Komnenovic V, Brownfield L, Twell D, Kakutani T, Tamaru H (2009) Induction of RNA-directed DNA methylation upon decondensation of constitutive heterochromatin. EMBO Rep 10(9):1015–1021
Tanaka I (1997) Differentiation of generative and vegetative cells in angiosperm pollen. Sex Plant Reprod 10:1–7
Tanaka I, Ono K, Fukuda T (1998) The developmental fate of angiosperm pollen is associated with a preferential decrease in the level of histone H1 in the vegetative nucleus. Planta 206:561–569
Terasaka O, Tanaka R (1974) Cytological studies on the nuclear differentiation in microspore division of some angiosperms. Bot Mag Tokyo 87:209–217
Ueda K, Kinoshita Y, Xu ZJ, Ide N, Ono M, Akahori Y, Tanaka I, Inoue M (2000) Unusual core histones specifically expressed in male gametic cells of Lilium longiflorum. Chromosoma 108:491–500
Vershinin AV, Alkhimova EG, HeslopHarrison JS (1996) Molecular diversification of tandemly organized DNA sequences and heterochromatic chromosome regions in some Triticeae species. Chromosome Res 4:517–525
Acknowledgements
The anti-CENH3 antibody was kindly provided by Drs. P. Talbert and S. Henikoff (Howard Hughes Medical Institute, USA). We thank Jochen Kumlehn and Ingo Schubert for helpful suggestions on the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (HO 1779/10-1 and SFB 648).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Hans de Jong.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. 1
Distribution of histone H3K4me2 in bicellular pollen of rye. Ten consecutive optical sections are shown. Note the clustering of immunosignals towards the periphery of the nuclear membrane and the exclusion of the nucleolus in the generative nucleus. In ‘merged’, DAPI and immunolabelled pollen are pseudocoloured in blue and in red, respectively. (PSD 6,281 kb; DOC 6,281 kb)
Fig. 2
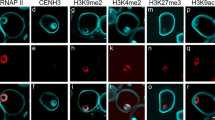
Distribution of a histone H3K4me2, b histone H3K9me2, c histone H3K9ac, d histone H3K27me3, e RNA polymerase II (RNA pol II) in meristematic root tip cells of rye. In ‘merged’, DAPI and immunolabelled cells are pseudocoloured in blue and in red, respectively. Scale bar represents 10 μm. (PSD 3,963 kb; DOC 3,963 kb)
Fig. 3
Distribution of CENH3 (in yellow) in meristematic metaphase chromosomes of rye. Chromosomes are counterstained in red. Scale bar represents 10 μm. (PSD 1,786 kb; DOC 1,786 kb)
Fig. 4
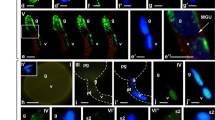
Distribution of CENH3 (in red) and of α-tubulin (in green) in tricellular pollen of rye. In ‘merged’, DAPI is shown in blue. Scale bar represents 50 μm. (PSD 7,188 kb; DOC 7,188 kb)
Fig. 5
Distribution of the centromere-specific repeat Bilby (in red) and of the subtelomere-specific repeat pSc200 (in green) in a semi-thin section of a rye anther. The insert in ‘merged’ shows part of the further enlarged filament. In ‘merged’, DAPI is shown in blue. Scale bar represents 100 μm. (PSD 7,254 kb; DOC 7,254 kb)
Rights and permissions
About this article
Cite this article
Houben, A., Kumke, K., Nagaki, K. et al. CENH3 distribution and differential chromatin modifications during pollen development in rye (Secale cereale L.). Chromosome Res 19, 471–480 (2011). https://doi.org/10.1007/s10577-011-9207-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-011-9207-6