Abstract
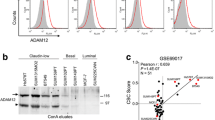
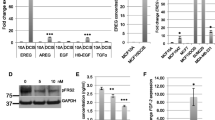
In the absence of HER2 overexpression, triple-negative breast cancers (TNBCs) rely on signaling by epidermal growth factor receptor (EGFR/ErbB1/HER1) to convey growth signals and stimulate cell proliferation. Soluble EGF-like ligands are derived from their transmembrane precursors by ADAM proteases, but the identity of the ADAM that is primarily responsible for ligand release and activation of EGFR in TNBCs is not clear. Using publicly available gene expression data for patients with lymph node-negative breast tumors who did not receive systemic treatment, we show that ADAM12L is the only ADAM with an expression level significantly associated with decreased distant metastasis-free survival times. Similar effect was not observed for patients with ER-negative non-TNBCs. There was a positive correlation between ADAM12L and HB-EGF and EGFR in TNBCs, but not in ER-negative non-TNBCs. We further demonstrate that ectopic expression of ADAM12L increased EGFR phosphorylation in a mouse intraductal xenograft model of early breast cancer. Finally, we detect strong correlation between the level of anti-ADAM12L and anti-phospho-EGFR immunostaining in human breast tumors using tissue microarrays. These studies suggest that ADAM12L is the primary protease responsible for the activation of EGFR in early stage, lymph node-negative TNBCs. Thus, our results may provide novel insight into the biology of TNBC.






Similar content being viewed by others
References
Carey L, Winer E, Viale G, Cameron D, Gianni L (2010) Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol 7(12):683–692. doi:10.1038/nrclinonc.2010.154
Foulkes WD, Smith IE, Reis-Filho JS (2010) Triple-negative breast cancer. New Engl J Med 363(20):1938–1948. doi:10.1056/NEJMra1001389
Pal SK, Childs BH, Pegram M (2011) Triple negative breast cancer: unmet medical needs. Breast Cancer Res Treat 125(3):627–636. doi:10.1007/s10549-010-1293-1
Sotiriou C, Pusztai L (2009) Gene-expression signatures in breast cancer. New Engl J Med 360(8):790–800. doi:10.1056/NEJMra0801289
Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM (2010) Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 12(5):R68. doi:10.1186/bcr2635
Hynes NE, MacDonald G (2009) ErbB receptors and signaling pathways in cancer. Curr Opin Cell Biol 21(2):177–184. doi:10.1016/j.ceb.2008.12.010
Foley J, Nickerson NK, Nam S, Allen KT, Gilmore JL, Nephew KP, Riese DJ 2nd (2010) EGFR signaling in breast cancer: bad to the bone. Semin Cell Dev Biol 21(9):951–960. doi:10.1016/j.semcdb.2010.08.009
Muraoka-Cook RS, Feng SM, Strunk KE, Earp HS III (2008) ErbB4/HER4: role in mammary gland development, differentiation and growth inhibition. J Mammary Gland Biol Neoplasia 13(2):235–246. doi:10.1007/s10911-008-9080-x
Wilson KJ, Gilmore JL, Foley J, Lemmon MA, Riese DJ II (2009) Functional selectivity of EGF family peptide growth factors: implications for cancer. Pharmacol Ther 122(1):1–8. doi:10.1016/j.pharmthera.2008.11.008
Horiuchi K, Le Gall S, Schulte M, Yamaguchi T, Reiss K, Murphy G, Toyama Y, Hartmann D, Saftig P, Blobel CP (2007) Substrate selectivity of epidermal growth factor-receptor ligand sheddases and their regulation by phorbol esters and calcium influx. Mol Biol Cell 18(1):176–188. doi:10.1091/mbc.E06-01-0014
Sunnarborg SW, Hinkle CL, Stevenson M, Russell WE, Raska CS, Peschon JJ, Castner BJ, Gerhart MJ, Paxton RJ, Black RA, Lee DC (2002) Tumor necrosis factor-α converting enzyme (TACE) regulates epidermal growth factor receptor ligand availability. J Biol Chem 277(15):12838–12845. doi:10.1074/jbc.M112050200
Sternlicht MD, Sunnarborg SW, Kouros-Mehr H, Yu Y, Lee DC, Werb Z (2005) Mammary ductal morphogenesis requires paracrine activation of stromal EGFR via ADAM17-dependent shedding of epithelial amphiregulin. Development 132(17):3923–3933. doi:10.1242/dev.01966
Sahin U, Weskamp G, Kelly K, Zhou HM, Higashiyama S, Peschon J, Hartmann D, Saftig P, Blobel CP (2004) Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J Cell Biol 164(5):769–779. doi:10.1083/jcb.200307137
Kenny PA, Bissell MJ (2007) Targeting TACE-dependent EGFR ligand shedding in breast cancer. J Clin Invest 117(2):337–345. doi:10.1172/JCI29518
McGowan PM, McKiernan E, Bolster F, Ryan BM, Hill AD, McDermott EW, Evoy D, O’Higgins N, Crown J, Duffy MJ (2008) ADAM-17 predicts adverse outcome in patients with breast cancer. Ann Oncol 19(6):1075–1081. doi:10.1093/annonc/mdm609
Edwards DR, Handsley MM, Pennington CJ (2008) The ADAM metalloproteinases. Mol Aspects Med 29(5):258–289. doi:10.1016/j.mam.2008.08.001
Kveiborg M, Albrechtsen R, Couchman JR, Wewer UM (2008) Cellular roles of ADAM12 in health and disease. Int J Biochem Cell Biol 40(9):1685–1702. doi:10.1016/j.biocel.2008.01.025
Kveiborg M, Frohlich C, Albrechtsen R, Tischler V, Dietrich N, Holck P, Kronqvist P, Rank F, Mercurio AM, Wewer UM (2005) A role for ADAM12 in breast tumor progression and stromal cell apoptosis. Cancer Res 65(11):4754–4761. doi:10.1158/0008-5472.CAN-05-0262
Frohlich C, Nehammer C, Albrechtsen R, Kronqvist P, Kveiborg M, Sehara-Fujisawa A, Mercurio AM, Wewer UM (2011) ADAM12 produced by tumor cells rather than stromal cells accelerates breast tumor progression. Mol Cancer Res 9(11):1449–1461. doi:10.1158/1541-7786.MCR-11-0100
Roy R, Rodig S, Bielenberg D, Zurakowski D, Moses MA (2011) ADAM12 transmembrane and secreted isoforms promote breast tumor growth: a distinct role for ADAM12-S protein in tumor metastasis. J Biol Chem 286(23):20758–20768. doi:10.1074/jbc.M110.216036
Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, Jatkoe T, Berns EM, Atkins D, Foekens JA (2005) Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365(9460):671–679. doi:10.1016/S0140-6736(05)17947-1
Minn AJ, Gupta GP, Padua D, Bos P, Nguyen DX, Nuyten D, Kreike B, Zhang Y, Wang Y, Ishwaran H, Foekens JA, van de Vijver M, Massague J (2007) Lung metastasis genes couple breast tumor size and metastatic spread. Proc Nat Acad Sci USA 104(16):6740–6745. doi:10.1073/pnas.0701138104
Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, Haibe-Kains B, Viale G, Delorenzi M, Zhang Y, d’Assignies MS, Bergh J, Lidereau R, Ellis P, Harris AL, Klijn JG, Foekens JA, Cardoso F, Piccart MJ, Buyse M, Sotiriou C (2007) Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res 13(11):3207–3214. doi:10.1158/1078-0432.CCR-06-2765
Schmidt M, Bohm D, von Torne C, Steiner E, Puhl A, Pilch H, Lehr HA, Hengstler JG, Kolbl H, Gehrmann M (2008) The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res 68(13):5405–5413. doi:10.1158/0008-5472.CAN-07-5206
Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA (2011) Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 121(7):2750–2767. doi:10.1172/JCI45014
Press MF, Finn RS, Cameron D, Di Leo A, Geyer CE, Villalobos IE, Santiago A, Guzman R, Gasparyan A, Ma Y, Danenberg K, Martin AM, Williams L, Oliva C, Stein S, Gagnon R, Arbushites M, Koehler MT (2008) HER-2 gene amplification, HER-2 and epidermal growth factor receptor mRNA and protein expression, and lapatinib efficacy in women with metastatic breast cancer. Clin Cancer Res 14(23):7861–7870. doi:10.1158/1078-0432.CCR-08-1056
Roepman P, Horlings HM, Krijgsman O, Kok M, Bueno-de-Mesquita JM, Bender R, Linn SC, Glas AM, van de Vijver MJ (2009) Microarray-based determination of estrogen receptor, progesterone receptor, and HER2 receptor status in breast cancer. Clin Cancer Res 15(22):7003–7011. doi:10.1158/1078-0432.CCR-09-0449
Cheadle C, Vawter MP, Freed WJ, Becker KG (2003) Analysis of microarray data using Z score transformation. J Mol Diagn 5(2):73–81. doi:10.1016/S1525-1578(10)60455-2
Behbod F, Kittrell FS, LaMarca H, Edwards D, Kerbawy S, Heestand JC, Young E, Mukhopadhyay P, Yeh HW, Allred DC, Hu M, Polyak K, Rosen JM, Medina D (2009) An intraductal human-in-mouse transplantation model mimics the subtypes of ductal carcinoma in situ. Breast Cancer Res 11(5):R66. doi:10.1186/bcr2358
Olayioye MA, Neve RM, Lane HA, Hynes NE (2000) The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J 19(13):3159–3167. doi:10.1093/emboj/19.13.3159
Cao Y, Kang Q, Zhao Z, Zolkiewska A (2002) Intracellular processing of metalloprotease disintegrin ADAM12. J Biol Chem 277(29):26403–26411. doi:10.1074/jbc.M110814200
Miller FR, Santner SJ, Tait L, Dawson PJ (2000) MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. J Natl Cancer Inst 92(14):1185–1186
Carey LA (2011) Directed therapy of subtypes of triple-negative breast cancer. Oncologist 16(Suppl 1):71–78. doi:10.1634/theoncologist.2011-S1-71
Shien T, Tashiro T, Omatsu M, Masuda T, Furuta K, Sato N, Akashi-Tanaka S, Uehara M, Iwamoto E, Kinoshita T, Fukutomi T, Tsuda H, Hasegawa T (2005) Frequent overexpression of epidermal growth factor receptor (EGFR) in mammary high grade ductal carcinomas with myoepithelial differentiation. J Clin Pathol 58(12):1299–1304. doi:10.1136/jcp.2005.026096
Arnes JB, Begin LR, Stefansson I, Brunet JS, Nielsen TO, Foulkes WD, Akslen LA (2009) Expression of epidermal growth factor receptor in relation to BRCA1 status, basal-like markers and prognosis in breast cancer. J Clin Pathol 62(2):139–146. doi:10.1136/jcp.2008.056291
Viale G, Rotmensz N, Maisonneuve P, Bottiglieri L, Montagna E, Luini A, Veronesi P, Intra M, Torrisi R, Cardillo A, Campagnoli E, Goldhirsch A, Colleoni M (2009) Invasive ductal carcinoma of the breast with the “triple-negative” phenotype: prognostic implications of EGFR immunoreactivity. Breast Cancer Res Treat 116(2):317–328. doi:10.1007/s10549-008-0206-z
Corkery B, Crown J, Clynes M, O’Donovan N (2009) Epidermal growth factor receptor as a potential therapeutic target in triple-negative breast cancer. Ann Oncol 20(5):862–867. doi:10.1093/annonc/mdn710
Sanchez-Munoz A, Gallego E, de Luque V, Perez-Rivas LG, Vicioso L, Ribelles N, Lozano J, Alba E (2010) Lack of evidence for KRAS oncogenic mutations in triple-negative breast cancer. BMC Cancer 10:136. doi:10.1186/1471-2407-10-136
Peddi PF, Ellis MJ, Ma C (2012) Molecular basis of triple negative breast cancer and implications for therapy. Int J Breast Cancer 2012:217185. doi:10.1155/2012/217185
Bhargava R, Gerald WL, Li AR, Pan Q, Lal P, Ladanyi M, Chen B (2005) EGFR gene amplification in breast cancer: correlation with epidermal growth factor receptor mRNA and protein expression and HER-2 status and absence of EGFR-activating mutations. Modern Pathol 18(8):1027–1033. doi:10.1038/modpathol.3800438
Reis-Filho JS, Pinheiro C, Lambros MB, Milanezi F, Carvalho S, Savage K, Simpson PT, Jones C, Swift S, Mackay A, Reis RM, Hornick JL, Pereira EM, Baltazar F, Fletcher CD, Ashworth A, Lakhani SR, Schmitt FC (2006) EGFR amplification and lack of activating mutations in metaplastic breast carcinomas. J Pathol 209(4):445–453. doi:10.1002/path.2004
Blobel CP, Carpenter G, Freeman M (2009) The role of protease activity in ErbB biology. Exp Cell Res 315(4):671–682. doi:10.1016/j.yexcr.2008.10.011
Duffy MJ, McKiernan E, O’Donovan N, McGowan PM (2009) Role of ADAMs in cancer formation and progression. Clin Cancer Res 15(4):1140–1144. doi:10.1158/1078-0432.CCR-08-1585
Duffy MJ, Mullooly M, O’Donovan N, Sukor S, Crown J, Pierce A, McGowan PM (2011) The ADAMs family of proteases: new biomarkers and therapeutic targets for cancer? Clin Proteomics 8(1):9. doi:10.1186/1559-0275-8-9
Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE (2006) The consensus coding sequences of human breast and colorectal cancers. Science 314(5797):268–274. doi:10.1126/science.1133427
Dyczynska E, Syta E, Sun D, Zolkiewska A (2008) Breast cancer-associated mutations in metalloprotease disintegrin ADAM12 interfere with the intracellular trafficking and processing of the protein. Int J Cancer 122(11):2634–2640. doi:10.1002/ijc.23405
Stautz D, Wewer UM, Kveiborg M (2012) Functional Analysis of a Breast Cancer-Associated Mutation in the Intracellular Domain of the Metalloprotease ADAM12. PLoS ONE 7(5):e37628. doi:10.1371/journal.pone.0037628
Lu Z, Jiang G, Blume-Jensen P, Hunter T (2001) Epidermal growth factor-induced tumor cell invasion and metastasis initiated by dephosphorylation and downregulation of focal adhesion kinase. Mol Cell Biol 21(12):4016–4031. doi:10.1128/MCB.21.12.4016-4031.2001
Lu Z, Ghosh S, Wang Z, Hunter T (2003) Downregulation of caveolin-1 function by EGF leads to the loss of E-cadherin, increased transcriptional activity of beta-catenin, and enhanced tumor cell invasion. Cancer Cell 4(6):499–515
Hager MH, Morley S, Bielenberg DR, Gao S, Morello M, Holcomb IN, Liu W, Mouneimne G, Demichelis F, Kim J, Solomon KR, Adam RM, Isaacs WB, Higgs HN, Vessella RL, Di Vizio D, Freeman MR (2012) DIAPH3 governs the cellular transition to the amoeboid tumour phenotype. EMBO Mol Med. doi:10.1002/emmm.201200242
Nie F, Yang J, Wen S, An YL, Ding J, Ju SH, Zhao Z, Chen HJ, Peng XG, Wong ST, Zhao H, Teng GJ (2012) Involvement of epidermal growth factor receptor overexpression in the promotion of breast cancer brain metastasis. Cancer. doi:10.1002/cncr.27553
Samanta S, Sharma VM, Khan A, Mercurio AM (2012) Regulation of IMP3 by EGFR signaling and repression by ERbeta: implications for triple-negative breast cancer. Oncogene. doi:10.1038/onc.2011.620
Jin W, Chen BB, Li JY, Zhu H, Huang M, Gu SM, Wang QQ, Chen JY, Yu S, Wu J, Shao ZM (2012) TIEG1 inhibits breast cancer invasion and metastasis by inhibition of epidermal growth factor receptor (EGFR) transcription and the EGFR signaling pathway. Mol Cell Biol 32(1):50–63. doi:10.1128/MCB.06152-11
Lu X, Wang Q, Hu G, Van Poznak C, Fleisher M, Reiss M, Massague J, Kang Y (2009) ADAMTS1 and MMP1 proteolytically engage EGF-like ligands in an osteolytic signaling cascade for bone metastasis. Genes Dev 23(16):1882–1894. doi:10.1101/gad.1824809
Fulford LG, Reis-Filho JS, Ryder K, Jones C, Gillett CE, Hanby A, Easton D, Lakhani SR (2007) Basal-like grade III invasive ductal carcinoma of the breast: patterns of metastasis and long-term survival. Breast Cancer Res 9(1):R4. doi:10.1186/bcr1636
Hicks DG, Short SM, Prescott NL, Tarr SM, Coleman KA, Yoder BJ, Crowe JP, Choueiri TK, Dawson AE, Budd GT, Tubbs RR, Casey G, Weil RJ (2006) Breast cancers with brain metastases are more likely to be estrogen receptor negative, express the basal cytokeratin CK5/6, and overexpress HER2 or EGFR. Am J Surg Pathol 30(9):1097–1104. doi:10.1097/01.pas.0000213306.05811.b9
Dent R, Hanna WM, Trudeau M, Rawlinson E, Sun P, Narod SA (2009) Pattern of metastatic spread in triple-negative breast cancer. Breast Cancer Res Treat 115(2):423–428. doi:10.1007/s10549-008-0086-2
Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massague J (2009) Genes that mediate breast cancer metastasis to the brain. Nature 459(7249):1005–1009. doi:10.1038/nature08021
McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM (2005) Reporting recommendations for tumor marker prognostic studies (REMARK). J Nat Cancer Inst 97(16):1180–1184. doi:10.1093/jnci/dji237
Acknowledgments
This work was supported by NIH grant 1R15CA151065 and Innovative Research Award from Terry C. Johnson Center for Basic Cancer Research at KSU to AZ, and by NIH grant 5R00CA127462 to FB. This is contribution 12-470-J from Kansas Agricultural Experiment Station.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Hui Li and Sara Duhachek-Muggy contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, H., Duhachek-Muggy, S., Qi, Y. et al. An essential role of metalloprotease-disintegrin ADAM12 in triple-negative breast cancer. Breast Cancer Res Treat 135, 759–769 (2012). https://doi.org/10.1007/s10549-012-2220-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-012-2220-4