Abstract
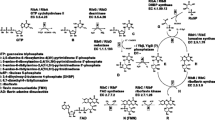
To study the network dynamics of the riboflavin biosynthesis pathway and to identify potential bottlenecks in the system, an ordinary differential equation-based model was constructed using available literature data for production strains. The results confirmed that the RibA protein is rate limiting in the pathway. Under the conditions investigated, we determined a potential limiting order of the remaining enzymes under increased RibA concentration (>0.102 mM) and therefore higher riboflavin production (>0.045 mmol g −1CDW h−1 and 0.0035 mM s−1, respectively). The reductase activity of RibG and lumazine synthase (RibH) might be the next most limiting steps. The computational minimization of the enzyme concentrations of the pathway suggested the need for a greater RibH concentration (0.251 mM) compared with the other enzymes (RibG: 0.188 mM, RibB: 0.023 mM).




Similar content being viewed by others
Abbreviations
- c :
-
Vector of metabolite concentrations
- t :
-
Simulation time, s
- N :
-
Stoichiometric matrix
- r :
-
Vector of reaction rates
- r 1 :
-
Reaction rate of the GTP cyclohydrolase II activity of RibA, mM s−1
- r 2 :
-
Reaction rate of the 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5′-phosphate deaminase activity of RibG, mM s−1
- r 3 :
-
Reaction rate of the 5-amino-6-ribosylamino-2,4(1H,3H)-pyrimidinedione 5′-phosphate reductase activity of RibG, mM s−1
- r 4 :
-
Reaction rate of the hypothetical phosphatase, mM s−1
- r 5 :
-
Reaction rate of the 3,4-dihydroxy-2-butanone 4-phosphate (DHBP) synthase activity of RibA, mM s−1
- r 6 :
-
Reaction rate of the 6,7-dimethyl-8-ribityllumazine synthase (RibH), mM s−1
- r 7 :
-
Reaction rate of the riboflavin synthase (RibB), mM s−1
- r 8 :
-
Rate of the diffusive outflow of riboflavin, mM s−1
- J 3 :
-
Reaction rate of the 5-amino-6-ribosylamino-2,4(1H,3H)-pyrimidinedione 5′-phosphate reductase activity of RibG at steady state, mM s−1
- J 6 :
-
Reaction rate of the 6,7-dimethyl-8-ribityllumazine synthase (RibH) at steady state, mM s−1
- J 7 :
-
Reaction rate of the riboflavin synthase (RibB) at steady state, mM s−1
- k cat :
-
Turnover number, s−1
- K m :
-
Michaelis–Menten constant, mM
- E RibA :
-
Enzyme concentration of RibA, mM
- E RibG :
-
Enzyme concentration of RibG, mM
- E RibH :
-
Enzyme concentration of RibH, mM
- E RibB :
-
Enzyme concentration of RibB, mM
- E RibX,min :
-
Minimized enzyme concentration of a specific Rib protein X, mM
- \( e_{\left[ A \right]}^{{r_{1} }} \) :
-
Scaled elasticity: sensitivity of the GTP cyclohydrolase II reaction rate to a change in the concentration of A
- \( e_{\left[ B \right]}^{{r_{2} }} \) :
-
Scaled elasticity: sensitivity of the deaminase reaction rate to a change in the concentration of B
- \( e_{\left[ C \right]}^{{r_{3} }} \) :
-
Scaled elasticity: sensitivity of the reductase reaction rate to a change in the concentration of C
- \( e_{\left[ F \right]}^{{r_{5} }} \) :
-
Scaled elasticity: sensitivity of the DHBP synthase reaction rate to a change in the concentration of F
- \( e_{\left[ E \right]}^{{r_{6} }} \) :
-
Scaled elasticity: sensitivity of the lumazine synthase reaction rate to a change in the concentration of E
- \( e_{\left[ G \right]}^{{r_{6} }} \) :
-
Scaled elasticity: sensitivity of the lumazine synthase reaction rate to a change in the concentration of G
- \( e_{\left[ H \right]}^{{r_{7} }} \) :
-
Scaled elasticity: sensitivity of the riboflavin synthase reaction rate to a change in the concentration of H
- μ :
-
Specific growth rate, s−1
- A :
-
GTP concentration, mM
- B :
-
2,5-Diamino-6-ribosylamino-4(3H)-pyrimidinone 5′-phosphate concentration, mM
- C :
-
5-Amino-6-ribosylamino-2,4(1H,3H)-pyrimidinedione 5′-phosphate concentration, mM
- D :
-
5-Amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione 5′-phosphate concentration, mM
- E :
-
5-Amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione concentration, mM
- F :
-
Ribulose 5-phosphate concentration, mM
- G :
-
3,4-Dihydroxy-2-butanone 4-phosphate (DHBP) concentration, mM
- H :
-
6,7-Dimethyl-8-ribityllumazine concentration, mM
- R :
-
Riboflavin concentration, mM
References
Abbas CA, Sibirny AA (2011) Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers. Microbiol Mol Biol Rev 75:321–360
Burgess CM, Smid EJ, van Sinderen D (2009) Bacterial vitamin B2, B11 and B12 overproduction: an overview. Int J Food Microbiol 133:1–7
Byrd RH, Hribar ME, Nocedal J (1998) An interior point algorithm for large scale nonlinear programming. SIAM J Optim 9:877–900
Dauner M, Sauer U (2001) Stoichiometric growth model for riboflavin-producing Bacillus subtilis. Biotechnol Bioeng 76:132–143
Echt S (2004) Biochemische und strukturelle Charakterisierung von Enzymen der Riboflavin-, Catecholamin- und Folsäurebiosynthese sowie des Calvin-Zyklus. Dissertation, Technical University Munich, Germany
Fischer M, Bacher A (2005) Biosynthesis of flavocoenzymes. Nat Prod Rep 22:324–350
Fischer M, Haase I, Kis K, Meining W, Ladenstein R, Cushman M, Schramek N, Huber R, Bacher A (2003) Enzyme catalysis via control of activation entropy: site-directed mutagenesis of 6,7-dimethyl-8-ribityl-lumazine synthase. J Mol Biol 326:783–793
Grima R, Schnell S (2008) Modelling reaction kinetics inside cells. Essays Biochem 45:41–56
Herz S, Eberhardt S, Bacher A (2000) Biosynthesis of riboflavin in plants. The ribA gene of Arabidopsis thaliana specifies a bifunctional GTP cyclohydrolase II/3,4-dihydroxy-2-butanone 4-phosphate synthase. Phytochemistry 53:723–731
Hohmann HP, Stahmann KP (2010) Biotechnology of riboflavin production. In: Mander L, Liu HW (eds) Comprehensive natural products, II chemistry and biology, vol 7., CofactorsElsevier, Philadelphia, pp 115–139
Hümbelin M, Griesser V, Keller T, Schurter W, Haiker M, Hohmann HP, Ritz H, Richter G, Bacher A, van Loon APGM (1999) GTP cyclohydrolase II and 3,4-dihydroxy-2-butanone 4-phosphate synthase are rate-limiting enzymes in riboflavin synthesis of an industrial Bacillus subtilis strain used for riboflavin production. J Ind Microbiol Biotechnol 22:1–7
Kato T, Park EY (2012) Riboflavin production by Ashbya gossypii. Biotechnol Lett 34:611–618
Kis K, Bacher A (1995) Substrate channeling in the lumazine synthase/riboflavin synthase complex of Bacillus subtilis. J Biol Chem 270:16788–16795
Klipp E, Heinrich R (1999) Competition for enzymes in metabolic pathways: implications for optimal distributions of enzyme concentrations and for the distribution of flux control. BioSystems 54:1–14
Klipp E, Liebermeister W, Wierling C, Kowald A, Lehrach H, Herwig R (2009) Systems biology: a textbook. Wiley, Weinheim
Lehmann M, Degen S, Hohmann HP, Wyss M, Bacher A, Schramek N (2009) Biosynthesis of riboflavin: screening for an improved GTP cyclohydrolase II mutant. FEBS J 276:4119–4129
Liebermeister W, Klipp E (2006) Bringing metabolic networks to life: convenience rate law and thermodynamic constraints. Theor Biol Med Model 3:41. doi:10.1186/1742-4682-3-41
Mack M, van Loon APGM, Hohmann HP (1998) Regulation of riboflavin biosynthesis in Bacillus subtilis is affected by the activity of the flavokinase/flavin adenine dinucleotide synthetase encoded by ribC. J Bacteriol 180:950–955
Magalhães MLB, Argyrou A, Cahill SM, Blanchard JS (2008) Kinetic and mechanistic analysis of the Escherichia coli ribD-encoded bifunctional deaminase–reductase involved in riboflavin biosynthesis. Biochemistry 47:6499–6507
Marx H, Mattanovich D, Sauer M (2008) Overexpression of the riboflavin biosynthetic pathway in Pichia pastoris. Microb Cell Fact 7:23. doi:10.1186/1475-2859-7-23
Moreno-Sánchez R, Saavedra E, Rodríguez-Enríquez S, Olín-Sandoval V (2008) Metabolic control analysis: a tool for designing strategies to manipulate metabolic pathways. J Biomed Biotechnol. doi:10.1155/2008/597913
Neidhardt FC, Ingraham JL, Schaechter M (1990) Physiology of the bacterial cell: a molecular approach. Sinauer Associates, Sunderland
Phillips R, Kondev J, Theriot J, Orme N (2008) Physical biology of the cell. Garland Science, New York
Richter G, Fischer M, Krieger C, Eberhardt S, Lüttgen H, Gerstenschläger I, Bacher A (1997) Biosynthesis of riboflavin: characterization of the bifunctional deaminase-reductase of Escherichia coli and Bacillus subtilis. J Bacteriol 179:2022–2028
Sauer U, Hatzimanikatis V, Bailey JE, Hochuli M, Szyperski T, Wüthrich K (1997) Metabolic fluxes in riboflavin-producing Bacillus subtilis. Nat Biotechnol 15:448–452
Schiffmann S (2002) Untersuchung von Strukturfunktionsbeziehungen bei Enzymen der Tetrahydrobioterin- und Riboflavinbiosynthese. Dissertation, Technical University Munich
Sengupta S, Kaufmann A, Chandra TS (2012) Development of fluorescent reporter tagged RIB gene cassettes for replicative transformation, early expression, and enhanced riboflavin production in Eremothecium ashbyi. Fungal Biol 116:1042–1051
Shampine LF (1994) Numerical solution of ordinary differential equations. Chapman & Hall, New York
Shi S, Shen Z, Chen X, Chen T, Zhao X (2009a) Increased production of riboflavin by metabolic engineering of the purine pathway in Bacillus subtilis. Biochem Eng J 46:28–33
Shi S, Chen T, Zhang Z, Chen X, Zhao X (2009b) Transcriptome analysis guided metabolic engineering of Bacillus subtilis for riboflavin production. Metab Eng 11:243–252
Stahmann KP, Revuelta JL, Seulberger H (2000) Three biotechnical processes using Ashbya gossypii, Candida famata or Bacillus subtilis compete with chemical riboflavin production. Appl Microbiol Biotechnol 53:509–516
Acknowledgments
This work was financed by the German Federal Ministry of Education and Research (BMBF) in the context of the NANOKAT Graduate Program (Ref. No. 0316052A). The study benefited from interdisciplinary discussions within the graduate program. The authors would especially like to thank Prof. Dr. M. Mack for his remarkable commitment within the program.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Birkenmeier, M., Neumann, S. & Röder, T. Kinetic modeling of riboflavin biosynthesis in Bacillus subtilis under production conditions. Biotechnol Lett 36, 919–928 (2014). https://doi.org/10.1007/s10529-013-1435-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-013-1435-8