Abstract
We conducted a meta-analysis to assess the association between tumor necrosis factor-alpha (TNF-alpha) gene TNFA-308 (G > A) and TNFA-857 (C > T) polymorphisms and gastric cancer (GC) susceptibility. We also performed subgroup analyses based on ethnicity (Caucasian, east Asian, and other populations) and tumor location [noncardia gastric cancer (NCGC)]. There were 3,335 GC patients and 5,286 controls for TNFA-308, and 1,118 GC patients and 1,591 controls for TNFA-857 in our analysis. Overall, allele contrast (A vs. G) of TNFA-308 polymorphism produced significant results in worldwide populations [P heterogeneity = 0.05, random-effects (RE) odds ratio (OR) 1.19; 95% confidence interval (CI) 1.03–1.37, P = 0.02] and Caucasian populations (P heterogeneity = 0.15, fixed-effects (FE), OR 1.27; 95% CI 1.11–1.45, P = 0.0005). Similar results were also obtained in recessive models and homozygote contrasts. No significant association was observed in NCGC and east Asian subgroup analysis. T variant of TNFA-857 produced significant results only in allele contrast (P heterogeneity = 0.38, FE OR 1.17; 95% CI 1.01–1.35, P = 0.04). In conclusion, TNFA-308 locus of TNF-alpha would be a risk factor for GC, especially in Caucasian populations. Besides, TNFA-857 locus may be related to GC risk, which demonstrated changeability of results in different contrasts.
Similar content being viewed by others
Introduction
Gastric cancer (GC) is now one of the most common cancers and a universal cause of death from cancer in the world. Studies suggest that GC is related to many factors, including Helicobacter pylori infection, gastroduodenal lesion, and genetic polymorphism, among which variants of pro- and anti-inflammatory cytokines such as interleukin (IL) and tumor necrosis factors (TNF) are most extensively investigated.
TNF-alpha is a proinflammatory cytokine initially identified as a serum factor inducing necrosis of transplanted tumors in mice (Carswell et al. 1975). It has been proven that TNF-alpha plays an essential role in host defense against infectious diseases, whereas its uncontrolled, excessive production may cause organ dysfunction (Strieter et al. 1993). There is also evidence suggesting that blood level of TNF-alpha is increased in solid tumors (Ardizzoia et al. 1992). Therefore, it seems likely that the expression level of TNF-alpha may be involved in cancer pathogenesis and progression. Functional reports of TNF-alpha also suggest that has an effect on GC risk, especially H. pylori-related GC. TNF-alpha is a potent gastric acid inhibitor (Beales and Calam 1998), and the inhibited acid secretion causes considerable change in H. pylori infection and its distribution on the corpus (Kuipers et al. 1996). It is well known that H. pylori infection increases the risk of gastric atrophy development and carcinogenesis.
The expression level of TNF-alpha is proven to be greatly influenced by polymorphisms in its promoter region (Raabe et al. 1998). Hence, polymorphisms in the TNFA gene promoter have been intensively studied as a putative determinant factor of susceptibility toward various diseases, including rheumatoid arthritis, psoriasis, and GC. There are many single nucleotide polymorphisms (SNPs) within the TNFA gene, such as TNFA-308 (G > A), TNFA-238 (G > A), TNFA-857 (C > T), and so on. Among them, the A allele of TNFA-308 polymorphism located in the promoter is associated with increased TNF-alpha production (Kroeger et al. 1997). Similarly, the T allele of TNFA-857 polymorphism located in the 5′-flanking region also shows higher transcriptional activity (Higuchi et al. 1998). Consequently, they are widely researched as potential susceptibility factors for GC.
In fact, there are numerous studies that focus on genetic polymorphisms and GC susceptibility or progression. However, the association between GC risk and polymorphisms found in TNFA is still controversial. El-Omar et al. found that proinflammatory genotypes of TNF-alpha were associated with more than doubling of the risk of noncardia GC (NCGC) in the USA (El-Omar et al. 2003). Similar conclusions were obtained by Machado in Portuguese and Hou in Polish populations (Hou et al. 2007; Machado et al. 2003). However, many other researchers’ results suggest that polymorphisms of TNFA may not be significantly associated with GC risk. These studies are based on limited sample size, and participant characteristics (such as tumor localization, ethnicity, age, gender, smoking status, and so on) are different from each other. Therefore, every single study may be underpowered to achieve a comprehensive and reliable conclusion.
To better address the association between TNFA-308 and TNFA-857 polymorphisms and GC risk, we performed a meta-analysis from all eligible studies, including subgroup analyses based on ethnicity and tumor localization.
Materials and methods
Study selection
All eligible studies were identified by searching the PubMed database for related reports published before November 2007 using the following search criterion: (“gastric” or “stomach”) and (“cancer” or “adenocarcinoma” or “carcinoma”) and (“polymorphism” or “polymorphisms”) and (“tumor necrosis factor-alpha” or “TNF-alpha” or “TNFA” or “TNF-A”). References of retrieved articles were also screened. Articles reporting the association between GC and TNFA-308 or TNFA-857 polymorphisms were identified. Included studies must meet all the following criteria: (a) based on unrelated individuals, pedigree data were excluded, (b) genotype distributions in both cases and controls were available, and (c) the genotype distribution of the control population must be in Hardy–Weinberg equilibrium (HWE).
Data extraction
Two investigators collected the data independently and reached a consensus on all items. The following basic information was extracted from the eligible studies: first author, year of publication, ethnicity of study population, sample size, type of sample for genotyping, tumor localization, gender, age, and other variables that can be sources of bias. Genotype distribution in both cases and controls were also obtained.
Statistical analysis
Meta-analysis
Odd ratios (OR) corresponding to a 95% confidence interval (CI) were used to assess the strength of association between TNFA polymorphisms and GC. Unadjusted OR of each study was first calculated in a 2 × 2 table. The meta-analysis examined the association between TNFA-308 allele A and GC risk compared with that for allele G (A vs. G); homozygote AA was contrasted with GG. Recessive (AA vs. GA + GG) and dominant (AA + GA vs. GG) models for allele A were also used. The same contrasts were performed for allele T of the TNFA-857 polymorphism. Between-study heterogeneity was assessed by a chi-square-based Q statistic test according to Lau (Lau et al. 1997). Heterogeneity was considered significant when P < 0.10. A random-effects (RE) model using the DerSimonian and Laird method and a fixed-effects (FE) model using the Mantel–Haenszel method were usually used to pool the results. Two methods produced similar results in the absence of heterogeneity. The RE model was more appropriate when a significant heterogeneity was present. Both the FE and RE models were used in Review-Manager 4.2 software, and pooled OR under both models were presented for a clearer comparison. The significance of pooled OR was tested by Z test (P < 0.05 was considered significant).
Population stratification
For TNFA-308 contrasts, subgroup analyses according to ethnicity (Caucasians, east Asians, and others) and tumor location (NCGC) were performed to estimate OR in specific populations. The TNFA-857 comparisons were not stratified for subgroup analysis because of the limited number of studies.
Sensitivity analysis
Through sequential omission of individual studies under various contrasts in Caucasian, east Asian and NCGC subgroups, respectively, sensitivity analysis was performed.
Assessment of publication bias
Funnel plots were used to investigate publication bias, where standard error of log (OR) of each study was plotted against its OR. Funnel plot asymmetry was assessed by the method of Egger’s linear regression test, a linear regression approach to measure funnel plot asymmetry on the natural logarithm scale of the OR (Egger et al. 1997). The significance of the intercept was determined by the t test, as suggested by Egger, and a P value of <0.05 was considered significant.
Software used
Hardy–Weinberg equilibrium was tested by the chi-square test using a Web-based program (http://www.ihg.gsf.de/cgi-bin/hw/hwa1.pl); Review-Manager 4.2 and stata 8.0 were also used in our meta-analysis. All P values were two-sided.
Results
Included studies
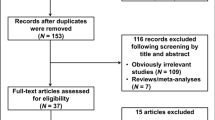
With our search criterion, 49 papers were found. Twenty-five papers concerning research on association between TNF-alpha and GC risk were identified, 21 of them meeting our criteria. Two papers (Fei et al. 2004; Seno et al. 2007) did not provide detailed genotype information. One paper (Wu et al. 2003) was excluded, as it was replaced by its later report (Wu et al. 2004). Three comparisons about TNFA-308 (Guo et al. 2005; Wu et al. 2004, 2002) and one comparison about TNFA-857 (Zambon et al. 2005) were excluded because the genotype distribution for the control population therein did not accord with HWE. Eighteen eligible articles (El-Omar et al. 2003; Garcia-Gonzalez et al. 2007, 2005; Glas et al. 2004; Hou et al. 2007; Jang et al. 2001; Kamangar et al. 2006; Kim et al. 2006; Lee et al. 2005, 2004; Li et al. 2005; Lu et al. 2005; Machado et al. 2003; Morgan et al. 2006; Perri et al. 2005; Rocha et al. 2005; Sugimoto et al. 2007; Zambon et al. 2005) referred to TNFA-308 polymorphism of TNFA; one of them had researched two geographic areas with different prevalence rates of GC, thus being treated as two independent studies (Perri et al. 2005). Five eligible articles (Li et al. 2005; Ohyama et al. 2004; Shirai et al. 2006; Sugimoto et al. 2007; Wu et al. 2004) were about TNFA-857 polymorphism.
Finally, 19 and five studies were considered for meta-analysis concerning TNFA-308 and TNFA-857, respectively. Populations were categorized into Caucasian, east Asian, mixed, and others. Data on NCGC patients were separated as a subgroup.
Meta-analysis database
Characteristics of the included studies are shown in Table 1. Among the 21 included articles, seven (33%) stated that controls were age-matched. All articles used blood samples for genotyping assay. Paraffin-embedded cancer tissue sections (two articles) and frozen biopsy specimens (three articles) were also used in some articles at the same time. A total of 3,335 GC patients and 5,286 controls (TNFA-308) and 1,118 GC patients and 1,591 controls (TNFA-857) were investigated. Genotype and allele distributions of TNFA -308 are shown in Table 2, including NCGC patient-based data. Nineteen comparisons (eight in Caucasians, seven in east Asians, two in the other population, two populations with mixed ethnic background) were considered for TNFA-308 and five comparisons (all in east Asians) for TNFA-857. The genotype GG was more common in east Asians (87.82% in patients and 88.44% in controls) than in Caucasians (70.86% in patients and 76.15% in controls), whereas GA (11.58% in patients and 11.14% in controls) and AA (0.60% in patients and 0.42% in controls) genotypes of this site were in a lower incidence in east Asians. Genotype and allele distributions of TNFA -857 are shown in Table 2.
Effects of allele and subgroup analysis
Pooled OR and other results are shown in Table 3. For TNFA -308 comparison, there was a significant between-study heterogeneity among 19 studies with respect to association between the A allele and GC susceptibility compared with the G allele (P heterogeneity = 0.05). Under the RE model, the pooled OR suggested that TNFA-308 allele A was associated with an increased risk of GC when compared with the G allele in worldwide populations (P = 0.02, RE OR 1.19; 95% CI 1.03–1.37, Fig. 1a). No significant heterogeneity existed in the subgroup analysis of Caucasian populations (P heterogeneity = 0.15). Under the FE model, a significant result was produced (A vs. G, FE OR 1.27; 95% CI 1.11–1.45, P = 0.0005). On the other hand, significant heterogeneity was observed in the subgroup analysis of east Asians (P heterogeneity = 0.04), and no significant association between the A allele and GC risk was detected (A vs. G, RE OR 1.06; 95% CI 0.72–1.56, P = 0.76). Since no AA genotype was detected in either the patient or control group in two east Asian population-based studies (Lee et al. 2005; Sugimoto et al. 2007) or in one study from Honduras (Morgan et al. 2006), OR of these studies could not be estimated under the recessive model and homozygote contrast. Under the recessive genetic model, AA genotype was associated with an increased risk of GC in worldwide populations (P heterogeneity = 0.82, FE OR 1.65; 95% CI 1.21–2.25, P = 0.001) and the Caucasian subgroup (P heterogeneity = 0.87, FE OR 1.55; 95% CI 1.01–2.36, P = 0.04) but not in east Asian and NCGC subgroups. Homozygote contrast also revealed similar effect: compared withTNFA-308 GG, significant association between TNFA-308 AA and the risk of GC existed in worldwide populations (P heterogeneity = 0.8, FE OR 1.78; 95% CI 1.28–2.48, P = 0.0007) and Caucasian populations (P heterogeneity = 0.86 FE OR 1.64; 95% CI 1.08–2.51, P = 0.02) (Table 3). In the NCGC patient-based subgroup study, no obvious associations were found between A allele or AA genotype and disease risk in all contrasts (Table 3).
Meta-analysis for tumor necrosis factor-alpha (TNFA)-308 polymorphism (A vs. G allele) in gastric cancer. The study is shown by a point estimate of the odds ratio (OR) and the accompanying 95% confidence interval (CI). n indicates the total number of A alleles; N indicates the total number of G plus A alleles. a Overall analysis in worldwide populations under a random-effects model. b Subgroup analysis in Caucasian populations under a fixed-effects model. c Subgroup analysis in east Asian populations under random-effects model. d Subgroup of noncardia gastric cancer (NCGC)-based cases under a fixed-effects model
A total of five studies were included in association investigation of TNFA-857 polymorphism and GC risk. No significant between-study heterogeneity was found in any comparison. There was a weak but significant association between the T allele and GC risk compared with the C allele (P heterogeneity = 0.38, FE OR 1.17; 95% CI 1.01–1.35, P = 0.04, Fig. 2). But other contrasts produced no significant results. Since all studies were in the east Asian population, no further segmentation could be made. Other relating characters of the meta-analysis are shown in Table 3.
Overall meta-analysis for tumor necrosis factor-alpha (TNFA)-857 polymorphism (T vs. C allele) in gastric cancer. The study is shown by a point estimate of the odds ratio (OR) and the accompanying 95% confidence interval (CI) under a fixed-effects model. n indicates the total number of T alleles; N indicates the total number of C plus T alleles
Sensitivity analysis
With regard to TNFA-308 polymorphism, the results pattern was not impacted by any single study in all subgroup studies (data not shown). Similar sensitivity analysis was also performed in comparisons of TNFA-857, the pooled OR (including 95% CI) consistently encompassed 1.0 in all contrasts when each single study referring to TNFA-857 polymorphism was excluded.
Publication bias
Funnel plot from comparisons of A versus G of TNFA-308 and T versus C of TNFA-857 polymorphism in the OR analysis was performed to assess publication bias. An Egger’s test was performed to statistically evaluate funnel plot symmetry, which provided no significant evidence for publication bias (t = -0.158, P = 0.132 for TNFA-308; t = 1.64, P = 0.20 for TNFA-857).
Discussion
This article investigated the relationship between TNFA polymorphisms and GC susceptibility. The overall results of this meta-analysis suggest that the A allele of the TNFA-308 locus causes an increased susceptibility to GC in worldwide and Caucasian populations. Comparisons under the recessive model and homozygote contrast supported the conclusion of allelic analysis. Sensitivity analysis showed that none of the results were affected by omission of any single study, indicating stability and reliability of the results. The association between TNFA-308 polymorphism and GC risk accorded with other functional research on TNFA, suggesting that the A allele of TNFA-308 site could apparently increase the transcription activity of this gene, and excessive expression of TNFA is related to inflammation or cancer promotion, as mentioned above. As shown in Table 3, no significant results were produced under the dominant model, implying that a dose effect may exist for this polymorphism.
However, such association was revealed to be insignificant in east Asian subgroup studies. Many factors may contribute to such discrepancy between Caucasian and east Asian populations. First, allele and genotype distribution of TNFA-308 locus was different in these two ethnicities. Percentage of A allele was especially low in east Asian populations, with 6.39% for cases and 5.99% for controls. Percentage of AA genotype was also lower in east Asian populations. Such statistical decrease might have led to the loss of association between TNFA-308 polymorphism and GC risk. Meanwhile, CIs for effect sizes in both Caucasian and east Asian populations overlapped with each other and suggest that there may be no differences between the two groups but simply a lack of power to detect an effect in east Asian studies. Moreover, different populations usually have different linkage disequilibrium (LD) patterns. For instance, a polymorphism locus may be in close linkage with another causal variant of disease risk. Such variant may exist only in a certain population and thus such association between the polymorphism and the disease risk may be lost in another population. Last but not least, clinical heterogeneity such as age, gender ratio, dietary, and disease severity of each population may also have an effect. Notably, the study by Garza-Gonzalez et al. ( 2005) in Mexico showed tremendously different genotype frequencies when compared with all other studies. And other analyses of TNFA-308 allele frequencies and genotype distribution in Mexican populations seem to be similar to those observed in other populations. Therefore, the reason for such difference in this study was unclear, and further research was needed.
No significant association between TNFA-308 polymorphism and NCGC was found. Discrepancy in GC and NCGC was also observed when researchers explored the association between H. pylori infection and GC risk (Huang et al. 1998). This may be attributed to different pathogenic mechanisms between NCGC and other kinds of GC. However, analysis with larger populations and contrasting analysis about both cardia GC and NCGC were desired to further explain the different effects of the A308 variant on them.
Our study also indicated that the T allele of TNFA-857 polymorphism may be associated with higher risk of GC in T vs. C comparison. The association was also supported by reports about increased transcriptional activity of the T allele and the pathological role of excessive expression of TNF-alpha. But analysis under dominant recessive models and homozygote contrast indicated insignificant association between TNFA-857 polymorphism and GC risk. Furthermore, results were easily affected by single study in sensitivity analysis, which indicated a limited number of studies and sample size, and current available studies were limited in east Asian populations. After reviewing additional studies on the association between TNFA-857 polymorphism and other diseases, we found that the percentage of T-allele carrier was slightly lower in Caucasian populations (data not shown). Theoretically, this would reduce the incidence of GC provided that the T allele was related to an increased risk of this disease. But explicit indication could not be made merely according to the genotype distribution of existing healthy controls. Other factors such as diet, genetic background, public sanitation, and so on must be considered at the same time. Hence, specific research on this polymorphism and GC risk was deemed necessary in Caucasian populations. Considering the fact that GC is a complex disease, many genetic variants may affect its pathogenesis and progression. Hence, haplotype analysis is recommended to explore the combined effects of polymorphisms.
H. pylori infection was also a modifier factor of GC. H. pylori strain and host genotypes possibly influenced the host inflammatory response and epithelial-cell physiology, thus changing the risk of carcinogenesis (Peek and Blaser 2002). Zambon et al. (2005) reported that H. pylori infection was associated with TNFA-308 genotype. Also, a meta-analysis suggested that H. pylori infection was strongly associated with early GC (Huang et al. 1998). Kim et al. (2006) provided separate genotyping data on H. pylori infection status-matched patients and healthy controls, which suggested that genetic polymorphisms could have considerable effect in H. pylori-induced gastroduodenal diseases. However, studies with detailed H. pylori infection status information were so limited that they were not sufficient to support analysis clarifying the modifying effect of H. pylori infection.
In conclusion, results of this meta-analysis indicate that the A allele of TNFA-308 polymorphism is related to increased incidence of GC, especially in Caucasian populations. TNFA-857 polymorphism T variant may be also correlated to higher GC risk. Further detailed investigation with larger numbers of worldwide participants is needed. And other confounding risk factors, such as age, gender, life style, and especially H. pylori infection status should also be taken into account when elucidating the clinically important risk factors for GC.
References
Ardizzoia A, Lissoni P, Brivio F, Tisi E, Perego MS, Grassi MG, Pittalis S, Crispino S, Barni S, Tancini G (1992) Tumor necrosis factor in solid tumors: increased blood levels in the metastatic disease. J Biol Regul Homeost Agents 6:103–107
Beales IL, Calam J (1998) Interleukin 1 beta and tumour necrosis factor alpha inhibit acid secretion in cultured rabbit parietal cells by multiple pathways. Gut 42:227–234
Carswell EA, Old LJ, Kassel RL, Green S, Fiore N, Williamson B (1975) An endotoxin-induced serum factor that causes necrosis of tumors. Proc Natl Acad Sci USA 72:3666–3670
Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629–634
El-Omar EM, Rabkin CS, Gammon MD, Vaughan TL, Risch HA, Schoenberg JB, Stanford JL, Mayne ST, Goedert J, Blot WJ, Fraumeni JF Jr, Chow WH (2003) Increased risk of noncardia gastric cancer associated with proinflammatory cytokine gene polymorphisms. Gastroenterology 124:1193–1201
Fei BY, Xia B, Deng CS, Xia XQ, Xie M, Crusius JB, Pena AS (2004) Association of tumor necrosis factor genetic polymorphism with chronic atrophic gastritis and gastric adenocarcinoma in Chinese Han population. World J Gastroenterol 10:1256–1261
Garza-Gonzalez E, Bosques-Padilla FJ, El-Omar E, Hold G, Tijerina-Menchaca R, Maldonado-Garza HJ, Perez-Perez GI (2005) Role of the polymorphic IL-1B, IL-1RN and TNF-A genes in distal gastric cancer in Mexico. Int J Cancer 114:237–241
Garcia-Gonzalez MA, Lanas A, Quintero E, Nicolas D, Parra-Blanco A, Strunk M, Benito R, Angel Simon M, Santolaria S, Sopena F, Piazuelo E, Jimenez P, Pascual C, Mas E, Irun P, Espinel J, Campo R, Manzano M, Geijo F, Pellise M, Gonzalez-Huix F, Nieto M, Espinos J, Tito L, Bujanda L, Zaballa M (2007) Gastric cancer susceptibility is not linked to pro-and anti-inflammatory cytokine gene polymorphisms in whites: a Nationwide Multicenter Study in Spain. Am J Gastroenterol 102:1878–1892
Glas J, Torok HP, Schneider A, Brunnler G, Kopp R, Albert ED, Stolte M, Folwaczny C (2004) Allele 2 of the interleukin-1 receptor antagonist gene is associated with early gastric cancer. J Clin Oncol 22:4746–4752
Guo W, Wang N, Li Y, Zhang JH (2005) Polymorphisms in tumor necrosis factor genes and susceptibility to esophageal squamous cell carcinoma and gastric cardiac adenocarcinoma in a population of high incidence region of North China. Chin Med J (Engl) 118:1870–1878
Higuchi T, Seki N, Kamizono S, Yamada A, Kimura A, Kato H, Itoh K (1998) Polymorphism of the 5′-flanking region of the human tumor necrosis factor (TNF)-alpha gene in Japanese. Tissue Antigens 51:605–612
Hou L, El-Omar EM, Chen J, Grillo P, Rabkin CS, Baccarelli A, Yeager M, Chanock SJ, Zatonski W, Sobin LH, Lissowska J, Fraumeni JF Jr, Chow WH (2007) Polymorphisms in Th1-type cell-mediated response genes and risk of gastric cancer. Carcinogenesis 28:118–123
Huang JQ, Sridhar S, Chen Y, Hunt RH (1998) Meta-analysis of the relationship between Helicobacter pylori seropositivity and gastric cancer. Gastroenterology 114:1169–1179
Jang WH, Yang YI, Yea SS, Lee YJ, Chun JH, Kim HI, Kim MS, Paik KH (2001) The -238 tumor necrosis factor-alpha promoter polymorphism is associated with decreased susceptibility to cancers. Cancer Lett 166:41–46
Kamangar F, Abnet CC, Hutchinson AA, Newschaffer CJ, Helzlsouer K, Shugart YY, Pietinen P, Dawsey SM, Albanes D, Virtamo J, Taylor PR (2006) Polymorphisms in inflammation-related genes and risk of gastric cancer (Finland). Cancer Causes Control 17:117–125
Kim N, Cho SI, Yim JY, Kim JM, Lee DH, Park JH, Kim JS, Jung HC, Song IS (2006) The effects of genetic polymorphisms of IL-1 and TNF-A on Helicobacter pylori-induced gastroduodenal diseases in Korea. Helicobacter 11:105–112
Kroeger KM, Carville KS, Abraham LJ (1997) The -308 tumor necrosis factor-alpha promoter polymorphism effects transcription. Mol Immunol 34:391–399
Kuipers EJ, Lundell L, Klinkenberg-Knol EC, Havu N, Festen HP, Liedman B, Lamers CB, Jansen JB, Dalenback J, Snel P, Nelis GF, Meuwissen SG (1996) Atrophic gastritis and Helicobacter pylori infection in patients with reflux esophagitis treated with omeprazole or fundoplication. N Engl J Med 334:1018–22
Lau J, Ioannidis JP, Schmid CH (1997) Quantitative synthesis in systematic reviews. Ann Intern Med 127:820–826
Lee SG, Kim B, Yook JH, Oh ST, Lee I, Song K (2004) TNF/LTA polymorphisms and risk for gastric cancer/duodenal ulcer in the Korean population. Cytokine 28:75–82
Lee JY, Kim HY, Kim KH, Kim SM, Jang MK, Park JY, Lee JH, Kim JH, Yoo JY (2005) Association of polymorphism of IL-10 and TNF-A genes with gastric cancer in Korea. Cancer Lett 225:207–214
Li C, Xia B, Yang Y, Li J, Xia HH (2005) TNF gene polymorphisms and Helicobacter pylori infection in gastric carcinogenesis in Chinese population. Am J Gastroenterol 100:290–294
Lu W, Pan K, Zhang L, Lin D, Miao X, You W (2005) Genetic polymorphisms of interleukin (IL)-1B, IL-1RN, IL-8, IL-10 and tumor necrosis factor {alpha} and risk of gastric cancer in a Chinese population. Carcinogenesis 26:631–636
Machado JC, Figueiredo C, Canedo P, Pharoah P, Carvalho R, Nabais S, Castro Alves C, Campos ML, Van Doorn LJ, Caldas C, Seruca R, Carneiro F, Sobrinho-Simoes M (2003) A proinflammatory genetic profile increases the risk for chronic atrophic gastritis and gastric carcinoma. Gastroenterology 125:364–371
Morgan DR, Dominguez RL, Keku TO, Heidt PE, Martin CF, Galanko JA, Omofoye OA, Sandler RS (2006) Gastric cancer and the high combination prevalence of host cytokine genotypes and Helicobacter pylori in Honduras. Clin Gastroenterol Hepatol 4:1103–11
Ohyama I, Ohmiya N, Niwa Y, Shirai K, Taguchi A, Itoh A, Hirooka Y, Wakai K, Hamajima N, Mori N, Goto H (2004) The association between tumour necrosis factor-alpha gene polymorphism and the susceptibility to rugal hyperplastic gastritis and gastric carcinoma. Eur J Gastroenterol Hepatol 16:693–700
Peek RM Jr, Blaser MJ (2002) Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer 2:28–37
Perri F, Piepoli A, Bonvicini C, Gentile A, Quitadamo M, Di Candia M, Cotugno R, Cattaneo F, Zagari MR, Ricciardiello L, Gennarelli M, Bazzoli F, Ranzani GN, Andriulli A (2005) Cytokine gene polymorphisms in gastric cancer patients from two Italian areas at high and low cancer prevalence. Cytokine 30:293–302
Raabe T, Bukrinsky M, Currie RA (1998) Relative contribution of transcription and translation to the induction of tumor necrosis factor-alpha by lipopolysaccharide. J Biol Chem 273:974–980
Rocha GA, Guerra JB, Rocha AM, Saraiva IE, da Silva DA, de Oliveira CA, Queiroz DM (2005) IL1RN polymorphic gene and cagA-positive status independently increase the risk of noncardia gastric carcinoma. Int J Cancer 115:678–683
Seno H, Satoh K, Tsuji S, Shiratsuchi T, Harada Y, Hamajima N, Sugano K, Kawano S, Chiba T (2007) Novel interleukin-4 and interleukin-1 receptor antagonist gene variations associated with non-cardia gastric cancer in Japan: comprehensive analysis of 207 polymorphisms of 11 cytokine genes. J Gastroenterol Hepatol 22:729–737
Shirai K, Ohmiya N, Taguchi A, Mabuchi N, Yatsuya H, Itoh A, Hirooka Y, Niwa Y, Mori N, Goto H (2006) Interleukin-8 gene polymorphism associated with susceptibility to non-cardia gastric carcinoma with microsatellite instability. J Gastroenterol Hepatol 21:1129–1135
Strieter RM, Kunkel SL, Bone RC (1993) Role of tumor necrosis factor-alpha in disease states and inflammation. Crit Care Med 21: S447–S463
Sugimoto M, Furuta T, Shirai N, Nakamura A, Xiao F, Kajimura M, Sugimura H, Hishida A (2007) Different effects of polymorphisms of tumor necrosis factor-alpha and interleukin-1 beta on development of peptic ulcer and gastric cancer. J Gastroenterol Hepatol 22:51–59
Wu MS, Huang SP, Chang YT, Shun CT, Chang MC, Lin MT, Wang HP, Lin JT (2002) Tumor necrosis factor-alpha and interleukin-10 promoter polymorphisms in Epstein-Barr virus-associated gastric carcinoma. J Infect Dis 185:106–109
Wu MS, Wu CY, Chen CJ, Lin MT, Shun CT, Lin JT (2003) Interleukin-10 genotypes associate with the risk of gastric carcinoma in Taiwanese Chinese. Int J Cancer 104:617–623
Wu MS, Chen LT, Shun CT, Huang SP, Chiu HM, Wang HP, Lin MT, Cheng AL, Lin JT (2004) Promoter polymorphisms of tumor necrosis factor-alpha are associated with risk of gastric mucosa-associated lymphoid tissue lymphoma. Int J Cancer 110:695–700
Zambon CF, Basso D, Navaglia F, Belluco C, Falda A, Fogar P, Greco E, Gallo N, Rugge M, Di Mario F, Plebani M (2005) Pro- and anti-inflammatory cytokines gene polymorphisms and Helicobacter pylori infection: interactions influence outcome. Cytokine 29:141–152
Acknowledgments
This work was supported by the National Science Foundation of China (30671145) and Program for New Century Excellent Talents in University (NCET).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Zhang, J., Dou, C., Song, Y. et al. Polymorphisms of tumor necrosis factor-alpha are associated with increased susceptibility to gastric cancer: a meta-analysis. J Hum Genet 53, 479–489 (2008). https://doi.org/10.1007/s10038-008-0273-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-008-0273-3
Keywords
This article is cited by
-
The Role of Tumor Necrosis Factor-α (TNF-α) Polymorphisms in Gastric Cancer: a Meta-Analysis
Journal of Gastrointestinal Cancer (2022)
-
Influence of functional polymorphisms in TNF-α, IL-8, and IL-10 cytokine genes on mRNA expression levels and risk of gastric cancer
Tumor Biology (2015)
-
Lack of association among TNF-α gene expression, -308 polymorphism (G > A) and virulence markers of Helicobacter pylori
Journal of Venomous Animals and Toxins including Tropical Diseases (2015)
-
IFRD1 polymorphisms and gastric cancer risk in a Chinese population
Medical Oncology (2014)
-
Association between TNF-α gene 308G>A polymorphism and lung cancer risk: a meta-analysis
Tumor Biology (2014)