Abstract
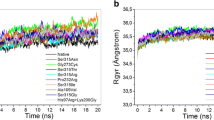
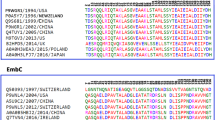
Remarkable advances have been made in the drug therapy of tuberculosis. However much remains to be learned about the molecular and structural basis of drug resistance in Mycobacterium tuberculosis. It is known that, activation of Isoniazid (INH) is mediated by Mycobacterium tuberculosis catalase-peroxidase (MtBKatG) and mutation at position 315 (serine to threonine) leads to resistance. We have conducted studies on the drug resistance through docking and binding analysis supported by time-scale (∼1000 ps) and unrestrained all-atom molecular dynamics simulations of wild and mutant MtBKatG. The study showed conformational changes of binding residues. Mutant (S315T) showed high docking score and INH binding affinity as compared to wild enzyme. In molecular dynamics simulation, mutant enzyme exhibited less structure fluctuation at INH binding residues and more degree of fluctuation at C-terminal domain compared to wild enzyme. Our computational studies and data endorse that MtBKatG mutation (S315T) decrease the flexibility of binding residues and made them rigid by altering the conformational changes, in turn it hampers the INH activity. We ascertain from this work that, this study on structural mechanism of resistance development in Mycobacterium tuberculosis would lead to new therapeutics based on the result obtained in this study.








Similar content being viewed by others
References
Zamocky M, Regelsberger G, Jakopitsch C, Obinger C (2001) The molecular peculiarities of catalase-peroxidases. FEBS Lett 492:177–182
Rouse DA, DeVito JA, Li Z, Byer H, Morris SL (1996) Site-directed mutagenesis of the katG gene of Mycobacterium tuberculosis: effects on catalase-peroxidase activities and isoniazid resistance. Mol Microbiol 22:583–592
Welinder KG (1991) Bacterial catalase-peroxidases are gene duplicated members of the plant peroxidase superfamily. Biochim Biophys Acta 1080:215–220
Nagy JM, Cass AE, Brown KA (1997) Purification and characterization of recombinant catalase-peroxidase, which confers isoniazid sensitivity in Mycobacterium tuberculosis. J Biol Chem 272:31265–31271
Zhang Y, Garbe T, Young D (1993) Transformation with katG restores isoniazid-sensitivity in Mycobacterium tuberculosis isolates resistant to a range of drug concentrations. Mol Microbiol 8:521–524
Singh AK, Kumar RP, Pandey N, Singh N, Sinha M, Bhushan A, Kaur P, Sharma S, Singh TP (2010) Mode of binding of the tuberculosis prodrug isoniazid to heme peroxidases: Binding studies and crystal structure of bovine lactoperoxidase with isoniazid at 2.7 A resolution. J Biol Chem 285:1569–1576
Wengenack NL, Rusnak F (2001) Evidence for isoniazid-dependent free radical generation catalyzed by Mycobacterium tuberculosis KatG and the isoniazid-resistant mutant KatG(S315T). Biochemistry 40:8990–8996
Timmins GS, Deretic V (2006) Mechanisms of action of isoniazid. Mol Microbiol 62:1220–1227
Johnsson K, King DS, Schultz PG (1995) Studies on the mechanism of action of isoniazid and ethionamide in the chemotherapy of tuberculosis. J Am Chem Soc 117:5009–5010
Hazbón MH, Brimacombe M, Bobadilla del Valle M, Cavatore M, Guerrero MI, Varma-Basil M, Billman-Jacobe H, Lavender C, Fyfe J, García-García L, León CI, Bose M, Chaves F, Murray M, Eisenach KD, Sifuentes-Osornio J, Cave MD, Ponce de León A, Alland D (2006) Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother 50:2640–2649
Marttila HJ, Mäkinen J, Marjamäki M, Soini H (2009) Prospective evaluation of pyrosequencing for the rapid detection of isoniazid and rifampin resistance in clinical Mycobacterium tuberculosis isolates. Eur J Clin Microbiol Infect Dis 28:33–38
Dalla Costa ER, Ribeiro MO, Silva MS, Arnold LS, Rostirolla DC, Cafrune PI, Espinoza RC, Palaci M, Telles MA, Ritacco V, Suffys PN, Lopes ML, Campelo CL, Miranda SS, Kremer K, da Silva PE, Fonseca L de S, Ho JL, Kritski AL, Rossetti ML (2009) Correlations of mutations in katG, oxyR-ahpC and inhA genes and in vitro susceptibility in Mycobacterium tuberculosis clinical strains segregated by spoligotype families from tuberculosis prevalent countries in South America. BMC Microbiol 9:39
Ng VH, Cox JS, Sousa AO, MacMicking JD, McKinney JD (2004) Role of KatG catalase-peroxidase in mycobacterial pathogenesis: Countering the phagocyte oxidative burst. Mol Microbiol 52:1291–1302
Marttila HJ, Soini H, Eerola E, Vyshnevskaya E, Vyshnevskiy BI, Otten TF, Vasilyef AV, Viljanen MK (1998) A Ser315Thr substitution KatG is predominant in genetically heterogeneous multidrug-resistant Mycobacterium tuberculosis isolates originating from the St. Petersburg area in Russia. Antimicrob Agents Chemother 42(9):2443–2445
Yu S, Girotto S, Lee C, Magliozzo RS (2003) Reduced affinity for Isoniazid in the S315T mutant of Mycobacterium tuberculosis KatG is a key factor in antibiotic resistance. J Biol Chem 278:14769–14775
Zhao X, Yu S, Ranguelova K, Suarez J, Metlitsky L, Schelvis JPM, Magliozzo RS (2009) Role of the oxyferrous heme intermediate and distal side adduct radical in the catalase activity of mycobacterium tuberculosis katg revealed by the w107f mutant. J Biol Chem 284:7030–7037
Ranguelova K, Suarez J, Metlitsky L, Yu S, Brejt SZ, Zhao L, Schelvis JP, Magliozzo RS (2008) Impact of distal side water and residue 315 on ligand binding to ferric Mycobacterium tuberculosis catalase-peroxidase (KatG). Biochemistry 47:12583–12592
Pym AS, Saint-Joanis B, Cole ST (2002) Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect Immun 70:4955–4960
Bertrand T, Eady NAJ, Jones JN, Jesmin NJM, Jamart-Gregoire B, Raven EL, Brown KA (2004) Crystal structure of Mycobacterium tuberculosis catalase-peroxidase. J Biol Chem 279:38991–38999
Heym B, Zhang Y, Poulet S, Young D, Cole ST (1993) Characterization of the katG gene encoding a catalase-peroxidase required for the isoniazid susceptibility of Mycobacterium tuberculosis. J Bacteriol 175:4255–4259
Zhao X, Yu H, Yu S, Wang F, Sacchettini JC, Magliozzo RS (2006) Hydrogen peroxide-mediated isoniazid activation catalyzed by Mycobacterium tuberculosis catalase-peroxidase (KatG) and its S315T mutant. Biochemistry 45:4131–4140
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucl Acids Res 28:235–242
Feldman J, Snyder KA, Ticoll A, Pintilie G, Hogue CW (2006) A complete small molecule dataset from the protein data bank. FEBS Lett 580:1649–1653
Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ (2005) PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res 33:363–367
Han LY, Lin HH, Li ZR, Zheng CJ, Cao ZW, Xie B, Chen YZ (2006) PEARLS: program for energetic analysis of receptor-ligand system. J Chem Inf Model 46(1):445–450
Chothia C, Janin J (1975) Principles of protein-protein recognition. Nature 256:705–708
Fraczkiewicz R, Braun W (1998) Exact and Efficient Analytical Calculation of the Accessible Surface Areas and Their Gradients for Macromolecules. J Comput Chem 19:319–333
Shrake A, Rupley JA (1973) Environment and exposure to solvent of protein atoms. Lysozyme and Insulin. J Mol Biol 79:351–371
Rodriguez R, Chinea G, Lopez N, Pons T (1998) Vriend G. Homology modeling, model and software evaluation: three related resources. Bioinformatics 14(6):523–528
Hess B, Kutzner C, Spoel D, Lindahl E (2008) GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447
Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ (2005) GROMACS: fast, flexible, and free. J Comput Chem 26:1701–1718
Van Gunsteren WF, Billeter SR, Eising AA, Hunenberger PH, Kruger P, Mark AE, Scott WRP, Tironi TG (1996) Biomolecular simulation: The Gromos 96 Manual and User Guide. Hochschulverlag AG an der Zurich, Zurich
Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J (1981) In: Pullman B (ed) Interaction models for water in relation to protein hydration. Intermolecular Forces. D Reidel Publishing Company, Dordrecht, pp 331–342
Berendsen HJC, Postma JP, van Gunsteren WF, DiNola A, Haak JR (1984) Molecular Dynamics with Coupling to an External bath. J Chem Phys 81:3684–3690
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh ewald method. J Chem Phys 103:8577–8593
Case DA, Pearlman DA, Caldwell JW, Wang J, Ross WS, Simmerling CL, Darden TA, Mertz KM, Stanton RV, Cheng AL, Vincent JJ, Crowley M, Tsue V, Gohlke H, Radmer R, Duan Y, Pitera J, Massova I, Seibel GL, Singh C, Weiner P, Kollman PA (2002) AMBER7 Simulation Software Package. University of California, San Francisco
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14:33–38
Emsley P, Cowtan K (2004) Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr D - Biol Cryst 60:2126–2132
Acknowledgments
We gratefully acknowledge the management of Vellore Institute of Technology University for providing the facilities to carry out this work. We thank the reviewers for their helpful comments and critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Purohit, R., Rajendran, V. & Sethumadhavan, R. Relationship between mutation of serine residue at 315th position in M. tuberculosis catalase-peroxidase enzyme and Isoniazid susceptibility: An in silico analysis. J Mol Model 17, 869–877 (2011). https://doi.org/10.1007/s00894-010-0785-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-010-0785-6