Abstract
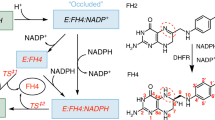
Dihydrofolate (DHF) reductase coded by a plasmid of the extremely halophilic archaeon Haloarcula japonica strain TR-1 (HjDHFR P1) shows moderate halophilicity on enzymatic activity at pH 6.0, although there is no significant effect of NaCl on its secondary structure. To elucidate the salt-activation and -inactivation mechanisms of this enzyme, we investigated the effects of pH and salt concentration, deuterium isotope effect, steady-state kinetics, and rapid-phase ligand-binding kinetics. Enzyme activity was increased eightfold by the addition of 500 mM NaCl at pH 6.0, fourfold by 250 mM at pH 8.0, and became independent of salt concentration at pH 10.0. Full isotope effects observed at pH 10.0 under 0–1000 mM NaCl indicated that the rate of hydride transfer, which was the rate-determining step at the basic pH region, was independent of salt concentration. Conversely, rapid-phase ligand-binding experiments showed that the amplitude of the DHF-binding reaction increased and the tetrahydrofolate (THF)-releasing rate decreased with increasing NaCl concentration. These results suggested that the salt-activation mechanism of HjDHFR P1 is via the population change of the anion-unbound and anion-bound conformers, which are binding-incompetent and -competent conformations for DHF, respectively, while that of salt inactivation is via deceleration of the THF-releasing rate, which is the rate-determining step at the neutral pH region.





Similar content being viewed by others
Abbreviations
- CD:
-
Circular dichroism
- DHF:
-
Dihydrofolate
- DHFR:
-
Dihydrofolate reductase
- EcDHFR:
-
DHFR from Escherichia coli
- HjDHFR:
-
DHFR from Haloarcula japonica
- HvDHFR:
-
DHFR from Haloferax volcanii
- MTE:
-
50 mM 2-(N-morpholino)ethanesulfonic acid, 25 mM Tris, and 25 mM ethanolamine containing 0.1 mM dithiothreitol and 0.1 mM EDTA (buffer)
- MTX:
-
Methotrexate
- NADPD:
-
4(R)-2H reduced nicotinamide adenine dinucleotide phosphate
- NADPH:
-
Nicotinamide adenine dinucleotide phosphate
- NMR:
-
Nuclear magnetic resonance
- THF:
-
Tetrahydrofolate
- TMACl:
-
Tetramethylammonium chloride
- TMAOH:
-
Tetramethylammonium hydroxide
References
Baccanari D, Phillips A, Smith S, Sinski D, Burchall J (1975) Purification and properties of Escherichia coli dihydrofolate reductase. Biochemistry 14:5267–5273
Behiry EM, Evans RM, Guo J, Loveridge EJ, Allemann RK (2014) Loop interactions during catalysis by dihydrofolate reductase from Moritella profunda. Biochemistry 53:4769–4774
Binbuga B, Boroujerdi AFB, Young JK (2007) Structure in an extreme environment: NMR at high salt. Protein Sci 16:1783–1787
Blecher O, Goldman S, Mevarech M (1993) High expression in Escherichia coli of the gene coding for dihydrofolate reductase of the extremely halophilic archae bacterium Haloferax volcanii. Eur J Biochem 216:199–203
Boroujerdi AFB, Young JK (2009) NMR-derived folate-bound structure of dihydrofolate reductase 1 from the halophile Haloferax volcanii. Biopolymers 91:140–144
Cayley PJ, Dunn SM, King RW (1981) Kinetics of substrate, coenzyme, and inhibitor binding to Escherichia coli dihydrofolate reductase. Biochemistry 20:874–879
Chen J, Taira K, Tu CD, Benkovic SJ (1987) Probing the functional role of phenylalanine-31 of Escherichia coli dihydrofolate reductase by site-directed mutagenesis. Biochemistry 26:4093–4100
David CL, Howell EE, Farnum MF, Villafranca JE, Oatley SJ, Kraut J (1992) Structure and function of alternative proton-relay mutants of dihydrofolate reductase. Biochemistry 31:9813–9822
Fierke CA, Johnson KA, Benkovic SJ (1987) Construction and evaluation of the kinetic scheme associated with dihydrofolate reductase from Escherichia coli. Biochemistry 26:4085–4092
Garvey EP, Matthews CR (1989) Effects of multiple replacements at a single position on the folding and stability of duhydrofolate reductase from Escherichia coli. Biochemistry 28:2083–2093
Gloss LM, Topping TB, Binder AK, Lohman JR (2008) Kinetic folding of Haloferax volcanii and Escherichia coli dihydrofolate reductases: Haloadaptation by unfolded state destabilization at high ionic strength. J Mol Biol 376:1451–1462
Grubbs J, Rahmanian S, DeLuca A, Padmashali C, Jackson M, Duff MR Jr, Howell E (2011) Thermodynamics and solvent effects on substrate and cofactor binding in Escherichia coli chromosomal dihydrofolate reductase. Biochemistry 50:3673–3685
Guo J, Luk LYP, Loveridge EJ, Allemann RK (2014) Thermal adaptation of dihydrofolate reductase from the moderate thermophile Geobacillus stearothermophilus. Biochemistry 53:2855–2863
Hamamoto T, Takashina T, Grant WD, Horikoshi K (1988) Asymmetric cell division of a triangular halophilic archaebacterium. FEMS Microbiol Lett 56:221–224
Horikoshi K, Aono R, Nakamura S (1993) The triangular halophilic archaebacterum Haloarcula japonica strain TR-1. Experientia 49:497–502
Jennings PA, Finn BE, Jones BE, Matthews CR (1993) A reexamination of the folding mechanism of dihydrofolate reductase from Escherichia coli: verification and refinement of a four-channel model. Biochemistry 32:3783–3789
Kuwajima K, Garvey EP, Finn BE, Matthews CR, Sugai S (1991) Transient intermediates in the folding of dihydrofolate reductase as detected by far-ultraviolet circular dichroism spectroscopy. Biochemistry 30:7693–7703
Luk LYP, Loveridge EJ, Allemann RK (2014) Different dynamical effects in mesophilic and hyperthermophilic dihydrofolate reductases. J Am Chem Soc 136:6862–6865
Miyashita Y, Ohmae E, Nakasone K, Katayanagi K (2015) Effects of salt on the structure, stability, and function of a halophilic dihydrofolate reductase from a hyper halophilic archaeon, Haloarcula japonica strain TR-1. Extremophiles 19:479–493
Nakamura S, Nakasone K, Takashina T (2010) Genetics and genomics of triangular disc-shaped halophilic archaeon Haloarcula japonica strain TR-1. In: Horikoshi K, Antranikian G, Bull A, Robb F, Stetter K (eds) Extremophiles Handbook. Springer, Tokyo, pp 363–381
Nishiyama Y, Takashina T, Grant WD, Horikoshi K (1992) Ultrastructure of the cell wall of the triangular halophilic archaebacterum Haloarcula japonica strain TR-1. FEMS Microbiol Lett 99:43–48
Ohmae E, Murakami C, Tate S, Gekko K, Hata K, Akasaka K, Kato C (2012) Pressure dependence of activity and stability of dihydrofolate reductases of the deep-sea bacterium Moritella profunda and Escherichia coli. Biochim Biophys Acta 1824:511–519
Ohmae E, Miyashita Y, Tate S, Gekko K, Kitazawa S, Kitahara R, Kuwajima K (2013) Solvent environments significantly affect the enzymatic function of Escherichia coli dihydrofolate reductase: comparison of wild-type protein and active-site mutant D27E. Biochim Biophys Acta 1834:2782–2794
Ortenberg R, Rozenblatt-Rosen O, Mevarech M (2000) The extremely halophilic archaeon Haloferax volcanii has two very different dihydrofolate reductases. Mol Microbiol 35:1493–1505
Osborne MJ, Venkitakrishnan RP, Dyson HJ, Wright PE (2003) Diagnostic chemical shift markers for loop conformation and substrate and cofactor binding in dihydrofolate reductase complexes. Protein Sci 12:2230–2238
Perry KM, Onuffer JJ, Touchette NA, Herndon CS, Gitteleman MS, Matthews CR, Chen JT, Mayer RJ, Taira K, Benkovic SJ, Howell EE, Kraut J (1987) Effects of single amino acid replacements on the folding and stability of dihydrofolate reductase from Escherichia coli. Biochemistry 26:2674–2682
Pieper U, Kapadia G, Mevarech M, Herzberg O (1998) Structural features of halophilicity derived from the crystal structure of dihydrofolate reductase from the Dead Sea halophilic archaeon, Haloferax volcanii. Structure 6:75–88
Roesser M, Müller V (2001) Osmoadaptation in bacteria and archaea: common principles and differences. Environ Microbiol 3:743–754
Sawaya MR, Kraut J (1997) Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence. Biochemistry 36:586–603
Stone SR, Morrison JF (1984) Catalytic mechanism of the dihydrofolate reductase reaction as determined by pH studies. Biochemistry 23:2753–2758
Viola RE, Cook PF, Cleland WW (1979) Stereoselective preperration of deuterated reduced nicotinamide adenine dinucleotide and substrates by enzymatic synthesis. Anal Biochem 96:334–340
Wang Z, Singh P, Czekster CM, Kohen A, Schramm VL (2014) Protein mass-modulated effects in the catalytic mechanism of dihydrofolate reductase: Beyond promoting vibration. J Am Chem Soc 136:8333–8341
Williams JW, Morrison JF, Duggleby RG (1979) Methotrexate, a high-affinity pseudosubstrate of dihydrofolate reductase. Biochemistry 18:2567–2573
Wright DB, Banks DD, Lohman JR, Hilsenbeck JL, Gloss LM (2002) The effects of salts on the activity and stability of Escherichia coli and Haloferax volcanii dihydrofolate reductases. J Mol Biol 323:327–344
Zusman T, Rosenshine I, Boehm G, Jaenicke R, Leskiw B, Mevarech M (1989) Dihydrofolate resuctase of the extremely halophilic archaebacterium Halobacterium volcanii. J Biol Chem 264:11878–11883
Acknowledgements
This work was supported financially by a Sasagawa Scientific Research Grant from the Japan Science Society (No. 26-315 to Y. M.) and the Platform Project for Supporting in Drug Discovery and Life Science Research (Platform for Dynamic Approaches to Living System) from Japan Agency for Medical Research and Development (AMED).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. Atomi.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Miyashita, Y., Ohmae, E., Ikura, T. et al. Halophilic mechanism of the enzymatic function of a moderately halophilic dihydrofolate reductase from Haloarcula japonica strain TR-1. Extremophiles 21, 591–602 (2017). https://doi.org/10.1007/s00792-017-0928-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-017-0928-0