Abstract
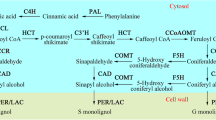
Tea plant, an economically important crop, is used in producing tea, which is a non-alcoholic beverage. Lignin, the second most abundant component of the cell wall, reduces the tenderness of tea leaves and affects tea quality. Caffeoyl-coenzyme A O-methyltransferase (CCoAOMT) involved in lignin biosynthesis affects the efficiency of lignin synthesis and lignin composition. A total of 10 CsCCoAOMTs were identified based on tea plant genome. Systematic analysis of CCoAOMTs was conducted for its physicochemical properties, phylogenetic relationships, conserved motifs, gene structure, and promoter cis-element prediction. Phylogenetic analysis suggested that all the CsCCoAOMT proteins can be categorized into three clades. The promoters of six CsCCoAOMT genes possessed lignin-specific cis-elements, indicating they are possibly essential for lignin biosynthesis. According to the distinct tempo-spatial expression profiles, five genes were substantially expressed in eight tested tissues. Most CsCCoAOMT genes were expressed in stems and leaves in three tea plant cultivars ‘Longjing 43,’ ‘Anjibaicha,’ and ‘Fudingdabai’ by RT-qPCR detection and analysis. The expression levels of two genes (CsCCoAOMT5 and CsCCoAOMT6) were higher than those of the other genes. The expression levels of most CsCCoAOMT genes in ‘Longjing 43’ were significantly higher than that those in ‘Anjibaicha’ and ‘Fudingdabai.’ Correlation analysis revealed that only the expression levels of CsCCoAOMT6 were positively correlated with lignin content in the leaves and stems. These results lay a foundation for the future exploration of the roles of CsCCoAOMTs in lignin biosynthesis in tea plant.







Similar content being viewed by others
References
Amiri ME, Asil MH (2007) Determination of optimum harvestable length of shoots in tea (Camellia sinensis L.) based on the current shoot growth, rather than interval plucking. J Food Agric Environ 5(2):122–124
Bailey TL, Boden M, Buske FA, Frith MC, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME suite: tools for motif discovery and searching. Nucleic Acids Res 37:202–208
Chen C, Meyermans H, Burggraeve B, De Rycke R, Inoue K, De Vleesschauwer V, Steenackers M, Van Montagu M, Engler G, Boerjan W (2000) Cell-specific and conditional expression of caffeoyl-coenzyme A-3-O-methyltransferase in poplar. Plant Physiol 123(3):853–867
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools—an integrative toolkit developed for interactive analyses of big biological data. bioRxiv:289660. https://doi.org/10.1101/289660
Day A, Neutelings G, Nolin F, Grec S, Habrant A, Cronier D, Maher B, Rolando C, David H, Chabbert B (2009) Caffeoyl coenzyme A O-methyltransferase down-regulation is associated with modifications in lignin and cell-wall architecture in flax secondary xylem. Plant Physiol Biochem 47(1):9–19
Deng W, Wang Y, Liu Z, Cheng H, Xue Y (2014) HemI: a toolkit for illustrating heatmaps. PLoS One 9(11):e111988
Do C, Pollet B, Thevenin J, Sibout R, Denoue D, Barriere Y, Lapierre C, Jouanin L (2007) Both caffeoyl coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta 226(5):1117–1129
Ferrer JL, Zubieta C, Dixon RA, Noel JP (2005) Crystal structures of Alfalfa caffeoyl coenzyme A 3-O-methyltransferase. Plant Physiol 137(3):1009–1017
Gao CY, Niu JG, Gao QC (2014) The application of origin software in the data processing of rock creep experiment. Appl Mech Mater 580-583:509–513
Giordano D, Provenzano S, Ferrandino A, Vitali M, Pagliarani C, Roman F, Cardinale F, Castellarin SD, Schubert A (2016) Characterization of a multifunctional caffeoyl-CoA O-methyltransferase activated in grape berries upon drought stress. Plant Physiol Biochem 101:23–32
Guo D, Chen F, Inoue K, Blount JW, Dixon RA (2001) Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic Alfalfa: impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13(1):73–88
Hamberger B, Ellis M, Friedmann M, Clarice DAS, Barbazuk B, Douglas CJ (2007) Genome-wide analyses of phenylpropanoid-related genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa: the Populus lignin toolbox and conservation and diversification of angiosperm gene families. Can J Bot 85(12):1182–1201
Hernandezgarcia CM, Finer JJ (2014) Identification and validation of promoters and cis-acting regulatory elements. Plant Sci 217:109–119
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27(1):297–300
Holt JS (2008) Book review: data analysis with SPSS: a first course in applied statistics. Teach Sociol 36(3):285–287
Hrazdina A, Jensen RA (1992) Spatial organization of enzymes in plant metabolic pathways. Annu Rev Plant Physiol Plant Mol Biol 43(1):241–267. https://doi.org/10.1146/annurev.pp.43.060192.001325
Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics (Oxford, England) 31(8):1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Hugueney P, Provenzano S, Verriès C, Ferrandino A, Meudec E, Batelli G, Merdinoglu D, Cheynier V, Schubert A, Ageorges A (2009) A novel cation-dependent O-methyltransferase involved in anthocyanin methylation in grapevine. Plant Physiol 150(4):2057–2070
Ibdah M, Zhang X, Schmidt J, Vogt T (2003) A novel Mg2+-dependent O-methyltransferase in the phenylpropanoid metabolism of Mesembryanthemum crystallinum. J Biol Chem 278(45):43961–43972
Inoue K, Sewalt VJH, Ballance GM, Ni W, Sturzer C, Dixon RA (1998) Developmental expression and substrate specificities of Alfalfa caffeic acid 3-O-methyltransferase and caffeoyl coenzyme A 3-O-methyltransferase in relation to lignification. Plant Physiol 117(3):761–770
Jia X, Wang G, Xiong F, Yu X, Xu Z, Wang F, Xiong A (2015) De novo assembly, transcriptome characterization, lignin accumulation, and anatomic characteristics: novel insights into lignin biosynthesis during celery leaf development. Sci Rep 5(1):8259–8259
Joshi CP, Chiang VL (1998) Conserved sequence motifs in plant S-adenosyl-L-methionine-dependent methyltransferases. Plant Mol Biol 37(4):663–674
Kai K, Mizutani M, Kawamura N, Yamamoto R, Tamai M, Yamaguchi H, Sakata K, Shimizu B (2008) Scopoletin is biosynthesized via ortho-hydroxylation of feruloyl CoA by a 2-oxoglutarate-dependent dioxygenase in Arabidopsis thaliana. Plant J 55(6):989–999
Kopycki JG, Rauh D, Chumanevich AA, Neumann P, Vogt T, Stubbs MT (2008) Biochemical and structural analysis of substrate promiscuity in plant Mg2+-dependent O-methyltransferases. J Mol Biol 378(1):154–164
Kumar S (2014) RNAi (RNA interference) vectors for functional genomics study in plants. Natl Acad Sci Lett 37(3):289–294
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Lacombe E, Van Doorsselaere J, Boerjan W, Boudet AM, Grimapettenati J (2000) Characterization of cis-elements required for vascular expression of the cinnamoyl CoA reductase gene and for protein–DNA complex formation. Plant J 23(5):663–676
Lam KCLC, Ibrahim RK, Behdad BB, Dayanandan S (2007) Structure, function, and evolution of plant O-methyltransferases. Genome 50(11):1001–1013
Lange H, Decina S, Crestini C (2013) Oxidative upgrade of lignin—recent routes reviewed. Eur Polym J 49(6):1151–1173
Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, Mcwilliam H, Valentin F, Wallace IM, Wilm A, Lopez R (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948
Lee YJ, Kim BG, Chong Y, Lim Y, Ahn J (2008) Cation dependent O-methyltransferases from rice. Planta 227(3):641–647
Leyva A, Liang X, Pintortoro JA, Dixon RA, Lamb CJ (1992) cis-Element combinations determine phenylalanine ammonia-lyase gene tissue-specific expression patterns. Plant Cell 4(3):263–271
Li L, Osakabe Y, Joshi CP, Chiang VL (1999) Secondary xylem-specific expression of caffeoyl-coenzyme A 3-O-methyltransferase plays an important role in the methylation pathway associated with lignin biosynthesis in loblolly pine. Plant Mol Biol 40(4):555–565
Li MY, Xu ZS, Tian C, Huang Y, Wang F, Xiong AS (2016) Genomic identification of WRKY transcription factors in carrot (Daucus carota) and analysis of evolution and homologous groups for plants. Sci Rep 6:23101
Li H, Liu Z, Wu Z, Wang Y, Teng R, Zhuang J (2018) Differentially expressed protein and gene analysis revealed the effects of temperature on changes in ascorbic acid metabolism in harvested tea leaves. Hortic Res 5(1):65
Liu S, Huang Y, Changjiu HE, Fang C, Zhang Y (2016a) Cloning, bioinformatics and transcriptional analysis of caffeoyl-coenzyme A 3-O-methyltransferase in switchgrass under abiotic stress. J Integr Agric 15(3):636–649
Liu X, Luo Y, Wu H, Xi W, Yu J, Zhang Q, Zhou Z (2016b) Systematic analysis of O-methyltransferase gene family and identification of potential members involved in the formation of O-methylated flavonoids in Citrus. Gene 575(2):458–472
Liu Z, Li H, Liu J, Wang Y, Zhuang J (2020) Integrative transcriptome, proteome, and microRNA analysis reveals the effects of nitrogen sufficiency and deficiency conditions on theanine metabolism in the tea plant (Camellia sinensis). Hortic Res 7(1):65. https://doi.org/10.1038/s41438-020-0290-8
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Lu J, Zhao H, Wei J, He Y, Shi C, Wang H, Song Y (2004) Lignin reduction in transgenic poplars by expressing antisense CCoAOMT gene. Prog Nat Sci 14(12):1060–1063
Lucker J, Martens S, Lund ST (2010) Characterization of a Vitis vinifera cv. Cabernet Sauvignon 3′, 5'-O-methyltransferase showing strong preference for anthocyanins and glycosylated flavonols. Phytochemistry 71(13):1474–1484
Meyermans H, Morreel K, Lapierre C, Pollet B, De Bruyn A, Busson R, Herdewijn P, Devreese B, Van Beeumen J, Marita JM (2000) Modifications in lignin and accumulation of phenolic glucosides in poplar xylem upon down-regulation of caffeoyl-coenzyme A O-methyltransferase, an enzyme involved in lignin biosynthesis. J Biol Chem 275(47):36899–36909
Moura JCMS, Bonine CAV, Viana JDOF, Dornelas MC, Mazzafera P (2010) Abiotic and biotic stresses and changes in the lignin content and composition in plants. J Integr Plant Biol 52(4):360–376
Nicholson RL, Hammerschmidt R (1992) Phenolic compounds and their role in disease resistance. Annu Rev Phytopathol 30(1):369–389
Patzlaff A, Mcinnis S, Courtenay A, Surman C, Newman LJ, Smith C, Bevan MW, Mansfield SD, Whetten RW, Sederoff RR (2003) Characterisation of a pine MYB that regulates lignification. Plant J 36(6):743–754
Proels RK, Oberhollenzer K, Pathuri IP, Hensel G, Kumlehn J, Huckelhoven R (2010) RBOHF2 of barley is required for normal development of penetration resistance to the parasitic fungus Blumeria graminis f. sp. hordei. Mol Plant-Microbe Interact 23(9):1143–1150
Que F, Wang G, Feng K, Xu Z, Wang F, Xiong A (2018) Hypoxia enhances lignification and affects the anatomical structure in hydroponic cultivation of carrot taproot. Plant Cell Rep 37(7):1021–1032
Raes J, Rohde A, Christensen JH, De Peer YV, Boerjan W (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133(3):1051–1071
Rakoczy M, Femiak I, Alejska M, Figlerowicz M, Podkowinski J (2018) Sorghum CCoAOMT and CCoAOMT-like gene evolution, structure, expression and the role of conserved amino acids in protein activity. Mol Gen Genomics 293(5):1077–1089
Rogers LA, Campbell MM (2004) The genetic control of lignin deposition during plant growth and development. New Phytol 164(1):17–30
Schmitt DL, An S (2017) Spatial organization of metabolic enzyme complexes in cells. Biochemistry 56(25):3184–3196
Shen J, Zou Z, Zhang X, Zhou L, Wang Y, Fang W, Zhu X (2018) Metabolic analyses reveal different mechanisms of leaf color change in two purple-leaf tea plant (Camellia sinensis L.) cultivars. Hortic Res 5(1):1–14
Shi J, Ma C, Qi D, Lv H, Yang T, Peng Q, Chen Z, Lin Z (2017) Erratum to: transcriptional responses and flavor volatiles biosynthesis in methyl jasmonate-treated tea leaves. BMC Plant Biol 17(1):136–136
Śmiechowska M, Dmowski P (2006) Crude fibre as a parameter in the quality evaluation of tea. Food Chem 94(3):366–368
Struck A, Thompson ML, Wong LS, Micklefield J (2012) S-adenosyl-methionine-dependent methyltransferases: highly versatile enzymes in biocatalysis, biosynthesis and other biotechnological applications. Chembiochem 13(18):2642–2655
Tamagnone L, Merida A, Parr AJ, Mackay S, Culianezmacia FA, Roberts K, Martin C (1998) The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10(2):135–154
Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W (2010) Lignin biosynthesis and structure. Plant Physiol 153(3):895–905
Vogt T (2004) Regiospecificity and kinetic properties of a plant natural product O-methyltransferase are determined by its N-terminal domain. FEBS Lett 561(1):159–162
Walker AM, Sattler SA, Regner M, Jones JP, Ralph J, Vermerris W, Sattler SE, Kang C (2016) The structure and catalytic mechanism of Sorghum bicolor caffeoyl-CoA O-methyltransferase. Plant Physiol 172(1):78–92
Wang W, Yang Y, Zhang W, Wu W (2014) Association of tea consumption and the risk of oral cancer: a meta-analysis. Oral Oncol 50(4):276–281
Wang Y, Wu X, Sun S, Xing G, Wang G, Que F, Khadr A, Feng K, Li T, Xu Z (2018) DcC4H and DcPER are important in dynamic changes of lignin content in carrot roots under elevated carbon dioxide stress. J Agric Food Chem 66(30):8209–8220
Wang Y, Teng R, Wang W, Wang Y, Shen W, Zhuang J (2019) Identification of genes revealed differential expression profiles and lignin accumulation during leaf and stem development in tea plant (Camellia sinensis (L.) O. Kuntze). Protoplasma 256(2):359–370
Wei JH, Zhao HY, Zhang JY, Liu HR, Song YR (2001) Cloning of cDNA encoding CCoAOMT from Populus tomentosa and downregulation of lignin content in transgenic plant expressing antisense gene. Acta Bot Sin 43(11):1179–1183
Wei K, Wang L, Zhang Y, Ruan L, Li H, Wu L, Xu L, Zhang C, Zhou X, Cheng H (2019) A coupled role for CsMYB75 and CsGSTF1 in anthocyanin hyperaccumulation in purple tea. Plant J 97(5):825–840
Weng J, Mo H, Chapple C (2010) Over-expression of F5H in COMT-deficient Arabidopsis leads to enrichment of an unusual lignin and disruption of pollen wall formation. Plant J 64(6):898–911
Widiez T, Hartman TG, Dudai N, Yan Q, Lawton M, Havkin-Frenkel D, Belanger FC (2011) Functional characterization of two new members of the caffeoyl CoA O-methyltransferase-like gene family from Vanilla planifolia reveals a new class of plastid-localized O-methyltransferases. Plant Mol Biol 76(6):475–488. https://doi.org/10.1007/s11103-011-9772-2
Wu ZJ, Tian C, Jiang Q, Li XH, Zhuang J (2016) Selection of suitable reference genes for qRT-PCR normalization during leaf development and hormonal stimuli in tea plant (Camellia sinensis). Sci Rep 6(1):19748
Xia E, Li F, Tong W, Li P, Wu Q, Zhao H, Ge R, Li R, Li Y, Zhang Z (2019) Tea plant information archive: a comprehensive genomics and bioinformatics platform for tea plant. Plant Biotechnol J 17(10):1938–1953
Xu Z, Zhang D, Hu J, Zhou X, Ye X, Reichel KL, Stewart NR, Syrenne RD, Yang X, Gao P (2009) Comparative genome analysis of lignin biosynthesis gene families across the plant kingdom. BMC Bioinformatics 10(11):1–15
Zhang J, Erickson L (2012) Harvest-inducibility of the promoter of alfalfa S-adenosyl-l-methionine: trans-caffeoyl-CoA3-O-methyltransferase gene. Mol Biol Rep 39(3):2489–2495
Zhang Q, Cai M, Yu X, Wang L, Guo C, Ming R, Zhang J (2017) Transcriptome dynamics of Camellia sinensis in response to continuous salinity and drought stress. Tree Genet Genomes 13(4):78
Zhao H, Wei J, Jinyu Z, Liu H, Tai W, Song Y (2002) Lignin biosynthesis by suppression of two O-methyl-transferases. Chin Sci Bull 47(13):1092–1095
Acknowledgments
The research was supported by the National Natural Science Foundation of China (31870681) and the Priority Academic Program Development of Jiangsu Higher Education Institutions Project (PAPD). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
JZ and SJL initiated and designed the experiments. SJL, YZY, RMT, HL, and HL performed the experiments. SJL analyzed the data. JZ contributed reagents/materials/analysis tools. SJL wrote the article. JZ, RMT, and HL revised the paper. All authors read and approved the final article.
Corresponding author
Ethics declarations
This article does not contain any studies with human participants or animal performed by any of the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Peter Nick
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lin, SJ., Yang, YZ., Teng, RM. et al. Identification and expression analysis of caffeoyl-coenzyme A O-methyltransferase family genes related to lignin biosynthesis in tea plant (Camellia sinensis). Protoplasma 258, 115–127 (2021). https://doi.org/10.1007/s00709-020-01555-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-020-01555-4