Abstract
Objective
In this study, we investigated the occurrence of papillomavirus (PV) infection in non-human primates (NHPs) in northeastern Argentina. We also explored their evolutionary history and evaluated the co-speciation hypothesis in the context of primate evolution.
Methods
We obtained DNA samples from 57 individuals belonging to wild and captive populations of Alouatta caraya, Sapajus nigritus, and Sapajus cay. We assessed PV infection by PCR amplification with the CUT primer system and sequencing of 337 bp (112 amino acids) of the L1 gene. The viral sequences were analyzed by phylogenetic and Bayesian coalescence methods to estimate the time to the most common recent ancestor (tMRCA) using BEAST, v1.4.8 software. We evaluated viral/host tree congruence with TreeMap v3.0.
Results
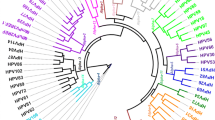
We identified two novel putative PV sequences of the genus Gammapapillomavirus in Sapajus spp. and Alouatta caraya (SPV1 and AcPV1, respectively). The tMRCA of SPV1 was estimated to be 11,941,682 years before present (ybp), and that of AcPV1 was 46,638,071 ybp, both before the coalescence times of their hosts (6.4 million years ago [MYA] and 6.8 MYA, respectively). Based on the comparison of primate and viral phylogenies, we found that the PV tree was no more congruent with the host tree than a random tree would be (P > 0.05), thus allowing us to reject the model of virus-host coevolution.
Conclusion
This study presents the first evidence of PV infection in platyrrhine species from Argentina, expands the range of described hosts for these viruses, and suggests new scenarios for their origin and dispersal.



Similar content being viewed by others
References
Rector A, Van Ranst M (2013) Animal papillomaviruses. Virology. https://doi.org/10.1016/j.virol.2013.05.007
de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H (2004) Classification of papillomaviruses. Virology. https://doi.org/10.1016/j.virol.2004.03.033
Van Doorslaer K, Li Z, Xirasagar S, Maes P, Kaminsky D, Liou D et al (2017) The papillomavirus episteme: a major update to the papillomavirus sequence database. Nucleic Acids Res. https://doi.org/10.1093/nar/gkw879
Antonsson A, Hansson BG (2002) Healthy skin of many animal species harbors papillomaviruses which are closely related to their human counterparts. J Virol. https://doi.org/10.1128/JVI.76.24.12537-12542.2002
Chen Z, van Doorslaer K, DeSalle R, Wood CE, Kaplan JR, Wagner JD et al (2009) Genomic diversity and interspecies host infection of alpha12 Macaca fascicularis papillomaviruses (MfPVs). Virology. https://doi.org/10.1016/j.virol.2009.07.012
Chen Z, Long T, Wong PY, Ho WCS, Burk RD, Chan PKS (2019) Non-human primate papillomaviruses share similar evolutionary histories and niche adaptation as the human counterparts. Front Microbiol. https://doi.org/10.3389/fmicb.2019.02093
Chan SY, Bernard HU, Ratterree M, Birkebak TA, Faras AJ, Ostrow RS (1997) Genomic diversity and evolution of papillomaviruses in rhesus monkeys. J Virol 71(7):4938–4943
Rector A, Lemey P, Tachezy R, Mostmans S, Ghim SJ, Van Doorslaer K et al (2007) Ancient papillomavirus-host co-speciation in Felidae. Genome Biol. https://doi.org/10.1186/gb-2007-8-4-r57
Gottschling M, Stamatakis A, Nindl I, Stockfleth E, Alonso A, Bravo IG (2007) Multiple evolutionary mechanisms drive papillomavirus diversification. Mol Biol Evol. https://doi.org/10.1093/molbev/msm039
Gottschling M, Göker M, Stamatakis A, Bininda-Emonds OR, Nindl I, Bravo IG (2011) Quantifying the phylodynamic forces driving papillomavirus evolution. Mol Biol Evol. https://doi.org/10.1093/molbev/msr030
Shah SD, Doorbar J, Goldstein RA (2010) Analysis of host-parasite incongruence in papillomavirus evolution using importance sampling. Mol Biol Evol. https://doi.org/10.1093/molbev/msq015
García-Pérez R, Ibáñez C, Godínez JM, Aréchiga N, Garin I, Pérez-Suárez G et al (2014) Novel papillomaviruses in free-ranging Iberian bats: no virus-host co-evolution, no strict host specificity, and hints for recombination. Genome Biol Evol. https://doi.org/10.1093/gbe/evt211
Bravo IG, de Sanjosé S, Gottschling M (2010) The clinical importance of understanding the evolution of papillomaviruses. Trends Microbiol. https://doi.org/10.1016/j.tim.2010.07.008
Van Doorslaer K (2013) Evolution of the papillomaviridae. Virology. https://doi.org/10.1016/j.virol.2013.05.012
Bolatti EM, Chouhy D, Casal PE, Pérez GR, Stella EJ, Sanchez A et al (2016) Characterization of novel human papillomavirus types 157, 158 and 205 from healthy skin and recombination analysis in genus γ-Papillomavirus. Infect Genet Evol. https://doi.org/10.1016/j.meegid.2016.04.018
Murahwa AT, Tshabalala M, Williamson AL (2020) Recombination between high-risk human papillomaviruses and non-human primate papillomaviruses: evidence of ancient host switching among alphapapillomaviruses. J Mol Evol. https://doi.org/10.1007/s00239-020-09946-0
Ostrow RS, McGlennen RC, Shaver MK, Kloster BE, Houser D, Faras AJ (1990) A rhesus monkey model for sexual transmission of a papillomavirus isolated from a squamous cell carcinoma. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.87.20.8170
Wood CE, Chen Z, Cline JM, Miller BE, Burk RD (2007) Characterization and experimental transmission of an oncogenic papillomavirus in female macaques. J Virol. https://doi.org/10.1128/JVI.00233-07
Bergin IL, Bell JD, Chen Z, Zochowski MK, Chai D, Schmidt K et al (2013) Novel genital alphapapillomaviruses in baboons (Papio hamadryas anubis) with cervical dysplasia. Vet Pathol. https://doi.org/10.1177/0300985812439725
Van Ranst M, Fuse A, Fiten P, Beuken E, Pfister H, Burk RD et al (1992) Human papillomavirus type 13 and pygmy chimpanzee papillomavirus type 1: comparison of the genome organizations. Virology. https://doi.org/10.1016/0042-6822(92)90896-W
Joh J, Hopper K, Van Doorslaer K, Sundberg JP, Jenson AB, Ghim SJ (2009) Macaca fascicularis papillomavirus type 1: a non-human primate betapapillomavirus causing rapidly progressive hand and foot papillomatosis. J Gen Virol. https://doi.org/10.1099/vir.0.006544-0
Wood CE, Tannehill-Gregg SH, Chen Z, Kv D, Nelson DR, Cline JM et al (2011) Novel betapapillomavirus associated with hand and foot papillomas in a cynomolgus macaque. Vet Pathol. https://doi.org/10.1177/0300985810383875
Chen Z, Wood CE, Abee CR, Burk RD (2018) Complete Genome sequences of three novel Saimiri sciureus papillomavirus types isolated from the cervicovaginal region of squirrel monkeys. Genome Announc. https://doi.org/10.1128/genomeA.01400-17
Long T, Wong PY, Ho WCS, Burk RD, Chan PKS, Chen Z (2018) Complete genome sequences of six novel Macaca mulatta papillomavirus types isolated from genital sites of Rhesus Monkeys in Hong Kong SAR, China. Microbiol Resour Announc. https://doi.org/10.1128/MRA.01414-18
Chen Z, DeSalle R, Schiffman M, Herrero R, Wood CE, Ruiz JC et al (2018) Niche adaptation and viral transmission of human papillomaviruses from archaic hominins to modern humans. PLoS Pathog. https://doi.org/10.1371/journal.ppat.1007352
Silvestre RV, de Souza AJ, Júnior EC, Silva AK, de Mello WA, Nunes MR et al (2016) First new world primate papillomavirus identification in the Atlantic Forest, Brazil: Alouatta guariba papillomavirus 1. Genome Announc. https://doi.org/10.1128/genomeA.00725-16
Köhler A, Gottschling M, Manning K, Lehmann MD, Schulz E, Krüger-Corcoran D et al (2011) Genomic characterization of ten novel cutaneous human papillomaviruses from keratotic lesions of immunosuppressed patients. J Gen Virol. https://doi.org/10.1099/vir.0.030593-0
Zunino GE, Kowalewski MM (2008) Primate research and conservation in northern Argentina: the field station Corrientes (Estación Biológica de Usos Múltiples—EBCo). Trop Conserv Sci. https://doi.org/10.1177/194008290800100206
Kowalewski MM, Salzer JS, Deutsch JC, Raño M, Kuhlenschmidt MS, Gillespie TR (2011) Black and gold howler monkeys (Alouatta caraya) as sentinels of ecosystem health: patterns of zoonotic protozoa infection relative to degree of human-primate contact. Am J Primatol. https://doi.org/10.1002/ajp.20803
International Primatological Society, 2014. Code of best practices for field primatology. https://www.asp.org/resources/docs/Code%20of_Best_Practices%20Oct%202014.pdf. Accessed 10 Jan 2022
Morales MA, Fabbri CM, Zunino GE, Kowalewski MM, Luppo VC, Enría DA et al (2017) Detection of the mosquito-borne flaviviruses, West Nile, Dengue, Saint Louis Encephalitis, Ilheus, Bussuquara, and Yellow Fever in free-ranging black howlers (Alouatta caraya) of Northeastern Argentina. PLoS Negl Trop Dis. https://doi.org/10.1371/journal.pntd.0005351
Nieves M, Remis MI, Sesarini C, Hassel DL, Argüelles CF, Mudry MD (2021) Assessment of genetic variability in captive capuchin monkeys (Primates: Cebidae). Sci Rep. https://doi.org/10.1038/s41598-021-86734-w
Chouhy D, Gorosito M, Sánchez A, Serra EC, Bergero A, Fernandez Bussy R, Giri AA (2010) New generic primer system targeting mucosal/genital and cutaneous human papillomaviruses leads to the characterization of HPV 115, a novel Beta-papillomavirus species 3. Virology. https://doi.org/10.1016/j.virol.2009.11.020
Bolatti EM, Hošnjak L, Chouhy D, Re-Louhau MF, Casal PE, Bottai H et al (2018) High prevalence of Gammapapillomaviruses (Gamma-PVs) in pre-malignant cutaneous lesions of immunocompetent individuals using a new broad-spectrum primer system, and identification of HPV210, a novel Gamma-PV type. Virology. https://doi.org/10.1016/j.virol.2018.09.006
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. https://doi.org/10.1093/nar/gkh340
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. https://doi.org/10.1038/nmeth.4285
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. https://doi.org/10.1093/molbev/msu300
Minh BQ, Nguyen MA, von Haeseler A (2013) Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. https://doi.org/10.1093/molbev/mst024
Rambaut, A. (2010) FigTree v1.3.1. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh. http://tree.bio.ed.ac.uk/software/figtree/. Accessed 10 Jan 2022.
Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A (2018) Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. https://doi.org/10.1093/ve/vey016
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior summarization in bayesian phylogenetics using tracer 1.7. Syst Biol. https://doi.org/10.1093/sysbio/syy032
Charleston (2011) TreeMap 3, which is freely available at https://sites.google.com/site/cophylogeny/software. Accessed 10 Jan 2022.
Page RD, Charleston MA (1997) From gene to organismal phylogeny: reconciled trees and the gene tree/species tree problem. Mol Phylogenet Evol. https://doi.org/10.1006/mpev.1996.0390
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. https://doi.org/10.1093/sysbio/syq010
D’arc M, Moreira FRR, Dias CA, Souza AR, Seuánez HN, Soares MA, Tavares MCH, Santos AFA (2020) The characterization of two novel neotropical primate papillomaviruses supports the ancient within-species diversity model. Virus Evol. https://doi.org/10.1093/ve/veaa036
Forslund O (2007) Genetic diversity of cutaneous human papillomaviruses. J Gen Virol. https://doi.org/10.1099/vir.0.82911-0
Bolatti EM, Hošnjak L, Chouhy D, Casal PE, Re-Louhau MF, Bottai H et al (2020) Assessing Gammapapillomavirus infections of mucosal epithelia with two broad-spectrum PCR protocols. BMC Infect Dis. https://doi.org/10.1186/s12879-020-4893-3
Bottalico D, Chen Z, Dunne A, Ostoloza J, McKinney S, Sun C et al (2011) The oral cavity contains abundant known and novel human papillomaviruses from the Betapapillomavirus and Gammapapillomavirus genera. J Infect Dis. https://doi.org/10.1093/infdis/jir383
Schino G, Di Giuseppe F, Visalberghi E (2009) Grooming, rank, and agonistic support in tufted capuchin monkeys. Am J Primatol. https://doi.org/10.1002/ajp.20627
Murahwa AT, Nindo F, Onywera H, Meiring TL, Martin DP, Williamson AL (2019) Evolutionary dynamics of ten novel Gamma-PVs: insights from phylogenetic incongruence, recombination and phylodynamic analyses. BMC Genom. https://doi.org/10.1186/s12864-019-5735-9
Perelman P, Johnson WE, Roos C, Seuánez HN, Horvath JE, Moreira MA et al (2011) A molecular phylogeny of living primates. PLoS Genet. https://doi.org/10.1371/journal.pgen.1001342
Lynch Alfaro JW, Boubli JP, Olson LE, Di Fiore A, Wilson B, Gutierrez-Espeleta GA et al (2011) Explosive pleistocene range expansion leads to widespread Amazonian sympatry between robust and gracile capuchin monkeys. J Biogeogr. https://doi.org/10.1111/j.1365-2699.2011.02609.x
Cortés-Ortiz L, Bermingham E, Rico C, Rodríguez-Luna E, Sampaio I, Ruiz-García M (2003) Molecular systematics and biogeography of the Neotropical monkey genus, Alouatta. Mol Phylogenet Evol. https://doi.org/10.1016/S1055-7903(02)00308-1
Williams JH, van Dyk E, Nel PJ, Lane E, Van Wilpe E, Bengis RG et al (2011) Pathology and immunohistochemistry of papillomavirus-associated cutaneous lesions in Cape mountain zebra, giraffe, sable antelope and African buffalo in South Africa. J S Afr Vet Assoc 82(3):185
Munday JS, Hanlon EM, Howe L, Squires RA, French AF (2007) Feline cutaneous viral papilloma associated with human papillomavirus type 9. Vet Pathol. https://doi.org/10.1354/vp.44-6-924
Arroyo LS, Smelov V, Bzhalava D, Eklund C, Hultin E, Dillner J (2013) Next generation sequencing for human papillomavirus genotyping. J Clin Virol. https://doi.org/10.1016/j.jcv.2013.07.013
Acknowledgements
We thank Maria Patricia Casco and Ester Bernaldo de Quirós for their help with sample collection during fieldwork. We are also thankful to María Elina Totaro for her technical assistance in the laboratory.
Funding
This work was supported through a doctoral fellowship from the Consejo Nacional de Investigaciones Científico y Tecnológicas (CONICET) (10320130101208CO) to CSF. IB, MMK, EB, DC, and AAG are members of CONICET. This study was partially supported by research Grant PIP IU 0355 CONICET (MMK). The equipment and software used in the study were partially supported by an Idea Wilds grant (CSF) (501c(3)) and a Codon Code Aligner Grants license program (IB). None of the funding agencies have been involved in the study design, data collection, analysis, or paper writing and submission.
Author information
Authors and Affiliations
Contributions
CS-F: conceptualization, methodology, validation, formal analysis, investigation, data curation, writing—original draft, writing—review and editing, visualization, funding acquisition. EMB: methodology, validation, formal analysis, investigation, data curation, writing—review and editing. ACAC: methodology, formal analysis, visualization, writing—review and editing. DC: methodology, formal analysis, writing—review and editing. MMK: conceptualization, resources, supervision, project administration, writing—review and editing. EJS: investigation, writing—review and editing. TGS: formal analysis, resources, writing—review and editing. MAR, DJL, and RHC: resources, writing—review and editing. AAG: resources; project administration, funding acquisition, writing—review and editing. IB: conceptualization, methodology, formal analysis, resources, data curation, writing—review and editing, supervision, project administration, funding acquisition
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Handling Editor: Graciela Andrei.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sanchez-Fernandez, C., Bolatti, E.M., Culasso, A.C.A. et al. Identification and evolutionary analysis of papillomavirus sequences in New World monkeys (genera Sapajus and Alouatta) from Argentina. Arch Virol 167, 1257–1268 (2022). https://doi.org/10.1007/s00705-022-05420-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-022-05420-y