Abstract
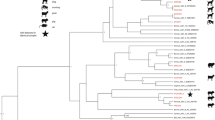
Adenoviruses have been identified in a wide variety of avian species, and in some species, they have been shown to cause disease and increase mortality. As part of an endeavor to investigate viruses associated with common terns (Sterna hirundo), a novel adenovirus was identified in fecal samples from two common terns on Gull Island, Lake Ontario, Canada. The coding-complete genome sequence of the new adenovirus is 31,094 bp, containing 28 putative genes, and this is the first adenovirus to be associated with terns. The virus was identified in two out of 13 fecal samples from tern chicks, and it was found to be most closely related to duck adenovirus 1, with the DNA polymerase sharing 58% amino acid sequence identity. Phylogenetic analysis based on DNA polymerase protein sequences showed that the new virus forms a distinct sub-branch within the atadenovirus clade and likely represents a new species in this genus.


Similar content being viewed by others
Data availability
The sequence described in this study has been deposited in the GenBank database under accession no. MW067004. The raw reads were deposited in SRA under project PRJNA780509; SRA accession no. SRR16948610.
References
Knowles DP (2017) Chapter 10—adenoviridae. In: MacLachlan NJ, Dubovi EJ (eds) Fenner’s veterinary virology, 5th edn. Academic Press, Boston, pp 217–227
Harrach B, Benkö M, Both GW, Brown M, Davison AJ, Echavarría M, Hess M, Kajon A, Lehmkuhl HD, Mautner V, Mittal SK, Wadell G (2012) Family—Adenoviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy. Elsevier, San Diego, pp 125–141
Salzmann E, Muller E, Marschang RE (2021) Detection of Testadenoviruses and Atadenoviruses in Tortoises and Turtles in Europe. J Zoo Wildl Med 52:223–231
Doszpoly A, Wellehan JF Jr, Childress AL, Tarjan ZL, Kovacs ER, Harrach B, Benko M (2013) Partial characterization of a new adenovirus lineage discovered in testudinoid turtles. Infect Genet Evol 17:106–112
Kajan GL, Kecskemeti S, Harrach B, Benko M (2013) Molecular typing of fowl adenoviruses, isolated in Hungary recently, reveals high diversity. Vet Microbiol 167:357–363
Del Valle FP, Camba SI, Umali DV, Sasai K, Shirota K, Katoh H, Tajima T (2020) Research note: molecular and pathologic characterization of avian adenovirus isolated from the oviducts of laying hens in eastern Japan. Poult Sci 99:2459–2468
Mukai Y, Tomita Y, Kryukov K, Nakagawa S, Ozawa M, Matsui T, Tomonaga K, Imanishi T, Kawaoka Y, Watanabe T, Horie M (2019) Identification of a distinct lineage of aviadenovirus from crane feces. Virus Genes 55:815–824
Kumar R, Sturos M, Porter RE, Singh A, Armien AG, Goyal SM, Mor SK (2021) An outbreak of Fowl aviadenovirus A-associated gizzard erosion and ulceration in captive bobwhite quail (Colinus virginianus). Avian Dis 65:52–58
Marek A, Kajan GL, Kosiol C, Harrach B, Schlotterer C, Hess M (2014) Complete genome sequences of pigeon adenovirus 1 and duck adenovirus 2 extend the number of species within the genus Aviadenovirus. Virology 462–463:107–114
Huang Y, Kang H, Dong J, Li L, Zhang J, Sun J, Zhang J, Sun M (2020) Isolation and partial genetic characterization of a new duck adenovirus in China. Vet Microbiol 247:108775
Zhang X, Zhong Y, Zhou Z, Liu Y, Zhang H, Chen F, Chen W, Xie Q (2016) Molecular characterization, phylogeny analysis and pathogenicity of a Muscovy duck adenovirus strain isolated in China in 2014. Virology 493:12–21
Teske L, Rubbenstroth D, Meixner M, Liere K, Bartels H, Rautenschlein S (2017) Identification of a novel aviadenovirus, designated pigeon adenovirus 2 in domestic pigeons (Columba livia). Virus Res 227:15–22
Kleine A, Hafez HM, Luschow D (2017) Investigations on aviadenoviruses isolated from turkey flocks in Germany. Avian Pathol 46:181–187
Marek A, Ballmann MZ, Kosiol C, Harrach B, Schlotterer C, Hess M (2014) Whole-genome sequences of two turkey adenovirus types reveal the existence of two unknown lineages that merit the establishment of novel species within the genus Aviadenovirus. J Gen Virol 95:156–170
Kajan GL, Davison AJ, Palya V, Harrach B, Benko M (2012) Genome sequence of a waterfowl aviadenovirus, goose adenovirus 4. J Gen Virol 93:2457–2465
Chen S, Cheng A, Wang M, Zhu D, Luo Q, Liu F, Chen X (2009) Detection and localization of a goose adenovirus in experimentally infected goslings, using indirect immunofluorescence with paraffin-embedded tissue sections. Avian Pathol 38:167–174
Oaks JL, Schrenzel M, Rideout B, Sandfort C (2005) Isolation and epidemiology of falcon adenovirus. J Clin Microbiol 43:3414–3420
Das S, Fearnside K, Sarker S, Forwood JK, Raidal SR (2017) A novel pathogenic aviadenovirus from red-bellied parrots (Poicephalus rufiventris) unveils deep recombination events among avian host lineages. Virology 502:188–197
Athukorala A, Forwood JK, Phalen DN, Sarker S (2020) Molecular characterisation of a novel and highly divergent passerine adenovirus 1. Viruses 12:1036
Gottdenker NL, Gregory CR, Ard MB, Lorenz WW, Nilsen RA, Ritchie BW (2019) Histopathologic changes, ultrastructure, and molecular characterization of an adenovirus in a sun conure (Aratinga solstitialis). Avian Dis 63:531–538
Marques MVR, Marin SY, Couto RM, Ecco R, Resende M, Martins N (2019) Fatal necrotic tracheitis by Aviadenovirus in captive Alagoas curassows (Pauxi mitu) extinct from the wild. Avian Pathol 48:278–283
Shah MS, Ashraf A, Khan MI, Rahman M, Habib M, Chughtai MI, Qureshi JA (2017) Fowl adenovirus: history, emergence, biology and development of a vaccine against hydropericardium syndrome. Arch Virol 162:1833–1843
Arnold JM, Oswald SA, Nisbet ICT, Pyle P, Patten MA (2020) Common Tern Sterna hirundo, version 1. In: Billerman M (ed) Birds of the world. Cornell Lab of Ornithology, Ithaca. https://doi.org/10.2173/bow.comter.01
Arnold JM, Tyerman DJ, Crump D, Williams KL, Oswald SA (2016) Premature feather loss among common tern chicks in Ontario: the return of an enigmatic developmental anomaly. PeerJ 4:1e1959
Arnold JM, Nisbet IC, Oswald SA (2016) Energetic constraint of non-monotonic mass change during offspring growth: a general hypothesis and application of a new tool. J Anim Ecol 85:476–486
Steel O, Kraberger S, Sikorski A, Young LM, Catchpole RJ, Stevens AJ, Ladley JJ, Coray DS, Stainton D, Dayaram A, Julian L, van Bysterveldt K, Varsani A (2016) Circular replication-associated protein encoding DNA viruses identified in the faecal matter of various animals in New Zealand. Infect Genet Evol 43:151–164
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Nurk S, Meleshko D, Korobeynikov A, Pevzner PA (2017) metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Bushnell B (2014) BBMap: a fast, accurate, splice-aware aligner. Lawrence Berkeley National Lab LBNL, Berkeley
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Darriba D, Taboada GL, Doallo R, Posada D (2011) ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165
Harrach B, Tarjan ZL, Benko M (2019) Adenoviruses across the animal kingdom: a walk in the zoo. FEBS Lett 593:3660–3673
Doszpoly A, Harrach B, LaPatra S, Benko M (2019) Unconventional gene arrangement and content revealed by full genome analysis of the white sturgeon adenovirus, the single member of the genus Ichtadenovirus. Infect Genet Evol 75:103976
Stover BC, Muller KF (2010) TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11:7
Acknowledgements
We thank Katie Rutt, Laura Ingraham, Anthony Mazza, Jenna Diehl, and Carolyn Degurski for assistance in the field. We also thank Christopher Kyle and Erica Nol for use of lab facilities at Trent University (Canada). Funding from Friends of Presqu’ile helped support fieldwork. B.H. was supported by the National Research, Development and Innovation Office, Hungary (NN140356). The molecular work was supported by Startup funds awarded to A.V. from Arizona State University (USA).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Ethical approval
All field study activities were approved by Penn State University’s Institutional Animal Care and Use Committee (protocol #45332) and permitted by the Canadian Wildlife Service Migratory Bird Act Scientific Permit CA 0308 and banding permit 10901 from Environment and Climate Change Canada.
Additional information
Handling Editor: Ana Cristina Bratanich.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kraberger, S., Oswald, S.A., Arnold, J.M. et al. Novel adenovirus associated with common tern (Sterna hirundo) chicks. Arch Virol 167, 659–663 (2022). https://doi.org/10.1007/s00705-021-05324-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05324-3