Abstract
Oral hypoglycemics are a widely prescribed group of drugs for the management of type 2 diabetes mellitus. Sulfonylureas (SUs) are the cornerstone of type 2 diabetes pharmacotherapy. The enzyme cytochrome P450 2C9 (CYP2C9) is the main enzyme that catalyzes the biotransformation of SUs. It is encoded by the polymorphic gene CYP2C9 with two allelic variants namely CYP2C9*2 and CYP2C9*3 coding for variant allozymes with reportedly decreased metabolic capacity resulting in decreased SUs clearance and consequently prolonged and exacerbated action. The aim of this study was to investigate the influence of genetic polymorphisms of CYP2C9 on the response to glibenclamide, a second-generation sulfonylurea. Hundred type 2 diabetic patients were enrolled in the study. Genotyping was done on the LightCycler 2 by real-time polymerase chain reaction (PCR) hybridization probe assay. The results are the following: 53 patients were carriers of the wild genotype (CYP2C9*1/*1), 20 were heterozygous for the variant CYP2C9*2 allele (CYP2C9*1/*2), 18 were heterozygous for the variant CYP2C9*3 allele (CYP2C9*1/*3), and 9 were double heterozygous for both mutant alleles (CYP2C9*2/*3); of those double mutant genotype patients, two were also homozygous for the mutant CYP2C9*2 allele (CYP2C9*2/*2) and one patient was homozygous for the mutant CYP2C9*3 allele (CYP2C9*3/*3). Although there was no significant difference in drug dosage between the four groups, there was however a significant association of the CYP2C9*2/*3 genotype with better glycemic control. As conclusion, the better glycemic control observed can probably be attributed to slower metabolism of SUs by the carriers of the CYP2C9*2/*3 genotype and consequently longer half-life or exacerbated effect of the SUs administered.
Similar content being viewed by others
Introduction
An estimated 9.1 % of the populations from the Middle Eastern/North African region have type 2 diabetes (32.8 million) in 2011, and this is projected to reach 60 million in 2030. Ranking prevalence of type 2 diabetes all over the world, Egypt comes seventh with a comparative prevalence of 16.9 % in the age group 20–79 years (IDF 2011).
Sulfonylureas (SUs) stimulate insulin release from pancreatic β-cells and have been the cornerstone of type 2 diabetes pharmacotherapy for over 50 years. First-generation SUs were less potent and required higher doses to achieve efficacy and are no longer used. While second-generation group including glyburide (glibenclamide), glipizide, and glimepiride are now widely used and are much more potent compounds (~100-fold); they possess a more rapid onset of action and generally have shorter plasma half-lives and longer duration of action compared to the first-generation agents (Evans and Rushakoff 2010).
Severe sulfonylurea-associated hypoglycemia is fatal in 1.4–10 % of cases and necessitates long and costly hospital stays (Holstein et al. 2005). Moreover, the risk of hypoglycemia may lead physicians to adjust blood glucose concentrations to above those identified as optimal for the prevention of microvascular, macrovascular, and neuropathic complications. Glibenclamide in particular has a greater tendency to cause hypoglycemic adverse effects than other sulfonylurea drugs (Nathan et al. 2009); thus, the identification and consideration of individual risk of hypoglycemia may be of great importance for the optimization of treatment.
When treating with SUs, one is bound to notice that there is considerable interindividual variability in pharmacokinetics, clinical efficacy, and adverse effects. Genetic factors are known to contribute to individual differences in bioavailability, drug transport, metabolism, and drug action. SUs are initiated at a low dose and titrated up to the optimal dose with intervals of 2–4 weeks until the glycemic target is achieved. Undertreatment will increase the risk of long-term complications, whereas overtreatment will lead to hypoglycemia (Kirchheiner et al. 2005).
Cytochrome P450 (CYP450) superfamily is a large and diverse group of enzymes. The function of most CYP enzymes is to catalyze the oxidation of organic substances. The substrates of CYP450 enzymes include metabolic intermediates such as lipids and steroid hormones, as well as drugs and other toxic chemicals. CYPs are the major enzymes involved in drug metabolism accounting for about 75 % of the total number of different metabolic reactions (Guengerich 2008).
Humans have 57 genes and more than 59 pseudogenes divided among 18 families of CYP450 genes and 43 subfamilies (Nelson 2002). Cytochrome P450 2C9 (abbreviated CYP2C9) is an enzyme that in humans is encoded by the CYP2C9 gene located within a cluster of cytochrome P450 genes on chromosome 10q24 (Inoue et al. 1994). It is an important cytochrome P450 enzyme with a major role in the oxidation of both xenobiotic and endogenous compounds.
Human CYP2C9 has been shown to exhibit genetic polymorphism. In addition to the wild type CYP2C9*1, at least 32 single nucleotide polymorphisms have been reported within the coding region of the CYP2C9 gene producing the variant allozymes. CYP2C9*2 and CYP2C9*3 are among them. CYP 29C*2 reflects an Arg144Cys (R144C) (King et al. 2004) and CYP2C9*3 reflects an Ile359-Leu (I359L) change in the amino acid sequence and have reduced catalytic activity compared with the wild type (CYP2C9*1) (Sullivan-Klose et al. 1996).
The enzyme CYP2C9 plays an important role in the pharmacokinetics of SUs, where the second-generation sulfonylureas including glibenclamide are extensively metabolized in the liver, mainly by CYP2C9 and the two allelic variants, CYP2C9*2 and CYP2C9*3, were reported by several authors to result in decreased metabolic capacity (Miners and Birkitt 1998; Kirchheiner et al. 2005; Zhou 2009). This in turn can result in increased plasma drug concentration and enhanced activity of the drug, which can manifest either as greater therapeutic response or a higher incidence of hypoglycemic episodes. Studies among healthy volunteers have shown a significant influence of CYP2C9 polymorphisms on glibenclamide metabolism (Kirchheiner et al. 2002; Niemi et al. 2002; Yin et al. 2005). There have also been reports of higher risk of hypoglycemic episodes in poor metabolizers of glibenclamide among diabetic patients (Holestein et al. 2005).
Aim of our work was to investigate the prevalence of the CYP2C9*2 and CYP2C9*3 genotypes in a cohort of type 2 diabetes patients on the sulfonylurea glibenclamide and the effect of different genotypes on treatment outcome.
Subjects and methods
Study design and patients selection
This study was conducted on a total number of 100 patients with type 2 diabetes in Kasr El Aini Diabetes Outpatient Clinic. Diabetes and diabetic complications were defined according to the American Diabetes Association criteria (2012). According to the Helsinki Declaration (World Medical Association 2008), the purpose of the study was explained to all the subjects included in the study and all participants gave their informed consent before joining the study and before physical examination and drawing of blood samples.
Inclusion criteria
Type 2 diabetes patients on SUs oral hypoglycemic therapy.
Clinical examination
Detailed diabetes and family history was accounted for; thorough medical examination was given to all participants including neurologic examination, blood pressure assessment, measurement of waist circumference, and calculation of body mass index.
Sampling
After an overnight fast, 5 ml blood sample was drawn from every patient. Two milliliters was collected on Na fluoride vacutainers for glucose assay and 3 ml on EDTA vacutainer for genotyping. Samples were transferred to laboratory where plasma was separated for glucose assay while EDTA samples were stored frozen for 2 weeks until processed (DNA extraction and genotyping).
Laboratory methods
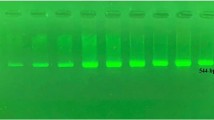
DNA extraction was done using the High Pure PCR Template Preparation Kit from Roche Diagnostics. Genotyping was done on Light Cycler 2 (Tortiello et al. 2006) using the LightMix human CYP2C9*2 and CYP2C9*3 kit (TIB MOLBIOL Syntheselabor GMBH, Eresburgstr 22–23 D-12103 Berlin).
Preparation of the reaction mix to the final volume of 20 μl consisted of 1.2 μl Mg+2 solution 25 mM, 4 μl reagent mix (primers and probes), 2 ml Roche master, and 5 μl of each sample that was completed to 20 μl by adding 7.8 μl water polymerase chain reaction (PCR)-grade.
The reaction consisted of amplification of a 374-bp fragment and a 180-bp fragment of the human CYP2C9 genome with specific primers. The CYP2C9*2 was detected by the SimpleProbe probe (detected in channel 530) while human CYP2C9*3 with probe labeled with lightcycler Red 640 (detected in channel 640). The genotypes were identified by running a melting curve with specific melting points. For CYP2C9*2, the melting temperature (T m) for the wild type was 58.5 °C and for the mutant 50.5 °C, detected in channel 530. For CYP2C9*3, the T m for the wild type was 48.3 °C and for the mutant 58.3 °C, detected in channel 640. The use of a color compensation file generated with the LightMix-Assay color Compensation 530/640 was employed to identify the genotypes in the corresponding channel.
The programmed protocol consisted of the following steps: denaturation step with sample denaturation and enzyme activation at 95 °C for 10 min; cycling of 45 cycles of PCR amplification of target DNA, at the following temperature profile, 95 °C for 5 s then at 60 °C for 10 s then 72 °C for 15 s; and finally melting curve analysis (Figs. 1, 2, and 3).
Glucose was assayed on automated analyzer Hitachi 917; commercial kits were supplied by (Roche Diagnostics, Germany) (Trinder 1969).
Statistical methods
The results were analyzed using the SPSS computer software package, version 15 (Chicago, IL, USA). Qualitative data were expressed as frequencies and percentages. For categorical data were assessed using chi-square test or Fischer exact test where appropriate. The quantitative data were expressed as median and range, and differences between the two groups were compared. Kruskal–Wallis test with Mann–Whitney U test for independent samples as post hoc multiple two-group comparisons was used. Logistic regression analysis was done to detect independent factors associated with blood sugar control. Differences were considered significant at P ≤ 0.05 (Knapp and Miller 1992).
Results
Of the 100 patients (19 males, 81 females), 94 were on concomitant treatment with two drugs, namely, glibenclamide and metformin, and six were on monotherapy with glibenclamide. There was no statistically significant difference in glibenclamide or metformin dose on comparing the various groups.
Table 1 shows genotype distribution among the hundred diabetic patients. The patients were divided into group 1, 53 patients carriers of the wild genotype designated CYP2C9*1/*1; group 2, 20 patients heterozygous for the variant genotype CYP2C9*2 designated CYP2C9*1/*2; group 3, 18 patients heterozygous carriers of the variant CYP2C9*3 genotype designated CYP2C9*1/*3; and group 4, nine patients heterozygous carriers of both variant genotypes CYP 2C9*2 and CYP2C9*3 designated CYP2C9*2/*3. In group 4, two patients were homozygous for the mutant CYP2C9*2 allele (CYP2C9*2/*2) as well as being heterozygous for the CYP2C9*3 and one patient was homozygous for the mutant CYP2C9*3 (CYP2C9*3/*3) allele as well as being heterozygous for the CYP2C9*2 allele, all those three patients were included in this group for statistical purposes.
Table 2 shows the characteristics of the four studied groups regarding age, gender, family history, fasting plasma glucose, duration of diabetes, BMI, waist circumference, hypertension, and history of ischemic heart disease. There was no statistically significant difference between the four studied groups regarding all those parameters.
On examining the association between the glibenclamide dosage and various parameters namely BMI, waist circumference, metformin dosage, neuropathy and other comorbidities, and control of fasting blood sugar in different genotypes, the following was found: there was a significant association (p = 0.015) between genotype status of CYP2C9 and the control of diabetes where only group 4 (double heterozygous) was associated with better diabetes control (FBS <130) (Table 3). There was no association between different genotypes and the rest of the studied parameters. By logistic regression analysis, we did not find significant interaction of the age, gender, BMI, waist circumference, family history, and duration of the treatment or comorbidities with control of fasting blood sugar or treatment efficacy on comparing the four groups.
Discussion
Comparing the prevalence of different CYP2C9 genotypes found in this study, we found 53 % of the patients homozygous for the wild type allele (CYP2C9*1/*1), 20 % heterozygous for the CYP2C9*2 allele, 18 % heterozygous for the CYP2C9*3 allele, and only 9 % double heterozygous (CYP2C9*2/*3) with two patients of this group also being homozygous for the mutant CYP2C9*2 allele and one homozygous for the mutant CYP2C9*3 allele.
Compared to the current study, the study by Holestein et al. (2005) revealed the following percentages in their three studied groups: a slightly higher prevalence of the wild genotype CYP2C9*1/*1 (65, 66.5, and 65 %, respectively), frequency very close to ours of the CYP2C9*1/*2 genotype (20, 18.4, and 19.6 %), lower frequency of CYP2C9*1/*3 genotype (5, 11.6, and 11.5 %), lower frequency of the CYP2C9*2/*3 (5, 1.5, and 1.7 %), and frequency very close to ours of CYP2C9*2/*2 genotype (0, 1.5, and 1.8 %) and for CYP2C9*3/*3 (5 %, 0.6 %, and 0.4 %).
Another study by Weise et al. (2010) that estimate the prevalence of CYP450 gene variations in 101 type 2 diabetes patients found the following: CYP2C9*2 genotype (20 %), CYP2C9*3 (22 %), and in 102 nondiabetics CYP2C9*2 (24 %) and CYP2C9*3 (21 %). This study shows close resemblance to the frequencies observed in our study.
Metformin (a biguanide) has a glucoregulatory effect only in the presence of endogenous insulin by decreasing endogenous glucose production and reducing peripheral resistance to insulin (approximately 20–30 %) (Campbell 2009), and used concomitantly with SUs, they are the most commonly prescribed medications for management of type 2 diabetic patients in the Kasr El Aini Diabetes outpatient Clinic. Indeed 94 of our patients were on combined metformin and glibenclamide therapy and only six were on monotherapy with glibenclamide. Metformin is not metabolized. It is cleared from the body by tubular secretion and excreted unchanged in the urine (Robert et al. 2003).
In the current study, there was no statistically significant difference regarding glibenclamide dosage in carriers of different genotypes, as well as no association of the dosage with genotype or metformin dosage or the other studied parameters. There was however an important observation, a significant association of the genotype with the diabetes control status taking the cutoff for controlled fasting plasma glucose of 130 mg/dl as recommended by the ADA (2011), where a better diabetes control was observed in carriers of the CYP2C9*2/*3 genotype. And since there was no significant difference in the metformin or glibenclamide dosage between the different studied groups, and since metformin is not metabolized by CYP450, we assume that the better control can be attributed to the different rate of metabolism of SUs administered, namely the slower metabolism by the carriers of the double heterozygous genotype (CYP2C9*2/*3). As early as 1998, Miners and Birkett observed that the genotypes CYP2C9*2/*3 and CYP2C9*3/*3 account for the slow metabolizer phenotype of many CYP2C9 substrates.
In the work by Kirchheiner in 2002, on healthy subjects, pharmacokinetics of glibenclamide depended significantly on CYP2C9 genotypes where in homozygous carriers of genotype CYP2C9*3/*3, total oral clearance was less than half that of the wild type genotype CYP2C9*1/*1. The study done by Niemi et al. (2002) on 29 healthy volunteers revealed that the CYP2C9*2/*3 genotype was associated with significantly lower clearance of SUs while the heterozygous genotypes with one wild type allele (CYP2C9*1/*3 and CYP2C9*1/*2) showed only a slight decrease in the oral clearances of glibenclamide.
Later work by Holestein et al. (2005) showed that sulfonylurea-treated patients who possessed the CYP2C9*3/*3 or CYP2C9*2/*3 genotype had 5.2-times the odds of a severe hypoglycemic events than the other CYP2C9 genotype groups and came to the conclusion that, although not the most prominent risk factor, CYP2C9 genotypes predictive of low enzyme activity (CYP2C9*3/*3 and CYP2C9*2/*3) should be considered as one but not the main risk factor for severe hypoglycemia resulting from treatment with sulfonylurea oral hypoglycemic agents.
Surendiran et al. (2011) came to the conclusion that genetic polymorphisms of CYP2C9 have a significant effect on the response to therapy with glibenclamide among 80 type 2 diabetes mellitus patients. In that study, the variant CYP2C9*1/*3 and CYP2C9*1/*2 genotypes were associated with controlled diabetic status, while the wild genotype of CYP2C9*1/*1 was associated with uncontrolled diabetic status; the cutoff for fasting blood sugar was 110 mg/dl.
On the other hand, the work done by Swen et al. (2010) to investigate the effect of CYP2C9 polymorphisms on prescribed dose and time-to-stable dose of sulfonylureas in primary care patients with type 2 diabetes mellitus came to the conclusion that no significant effects of the CYP2C9*2 and CYP2C9*3 alleles were found. However, a trend towards a lower stable SUs dose for carriers of the CYP2C9*3 allele was observed and it was their conclusion that genotyping for the CYP2C9*2 and CYP2C9*3 alleles currently appears to have no clinical implications for dosing of SUs in primary care patients with type 2 diabetes mellitus.
In conclusion, in the current study, since the glibenclamide drug dosage did not differ significantly among different genotypes, the association of CYP2C9*2/*3 genotype with better diabetes control can probably be attributed to slower metabolism and consequently longer half-life. The field of pharmacogenomics of oral antidiabetic medications has taken some big steps in recent years. Diabetes being the new world pandemic with its high prevalence and the ever expanding population of type 2 diabetes patients is entitled to an ongoing in-depth look into pharmacogenomics of various drugs used for its management, bearing in mind that better control means less short- and long-term complications and far less economic burden.
References
American Diabetes Association (2011) Standards of Medical Care in Diabetes—2011. Diabetes Care 34:S11–S61. doi:10.2337/dc11-S011
American Diabetes Association (2012) Diagnosis and classification of diabetes mellitus. Diabetes Care 35(Suppl 1):S64–S71
Campbell RK (2009) Type 2 diabetes: where we are today: an overview of disease burden, current treatments, and treatment strategies. J Am Pharm Assoc 49(Suppl 1):S3–S9. doi:10.1331/JAPhA.2009.09077
Evans JL and Rushakoff RJ. Oral pharmacological agents for type 2 diabetes (2010): oral agents, incretions and other “non-insulin” pharmacologic interventions for diabetes, Endo Text. Org.—The Endocrine Source, Diabetes Manager (Eds. I. D. Goldfine and R. J. Rushakoff) last author version May 2010; Available at http://diabetesmanager.pbworks.com/w/page/17680289/Oral-Pharmacological-Agents-for-Type-2-Diabetes. Accessed 31 July 2012
Guengerich FP (2008) Cytochrome p450 and chemical toxicology. Chem Res Toxicol 21(1):70–83
Holstein A, Plaschke A, Ptak M, Egberts EH, El-Din J, Brockmöller J, Kirchheiner J (2005) Association between CYP2C9 slow metabolizer genotypes and severe hypoglycaemia on medication with sulphonylurea hypoglycaemic agents. Br J Clin Pharmacol 60:103–106
Inoue K, Inazawa J, Suzuki Y, Shimada T, Yamazaki H, Guengerich PF, Abe T (1994) Fluorescencein situ hybridization analysis of chromosomal localization of three human cytochrome P450 2C genes (CYP2C8, 2C9, and 2C10) at 10q24.1. Jpn J Hum Genet 39(3):337–343
International Diabetes Federation. IDF (2011) Diabetes Atlas, 5th edn. International Diabetes Federation, Brussels. Available at: http://www.idf.org/diabetesatlas
King BP, Khan TI, Aithal GP, Kamali F, Daly AK (2004) Upstream and coding region CYP2C9 polymorphisms: correlation with warfarin dose and metabolism. Pharmacogenetics 14(12):813–822
Kirchheiner J, Brockmoller J, Meineke I, Bauer S, Rohde W, Meisel C et al (2002) Impact of CYP2C9 amino acid polymorphisms on glyburide kinetics and on the insulin and glucose response in healthy volunteers. Clin Pharmacol Ther 71:286–296
Kirchheiner J, Roots I, Rosenkranz B, Goldammer M, Brockmoller J (2005) Effect of genetic polymorphisms in cytochrome p450 (CYP) 2C9 andCYP2C8 on the pharmacokinetics of oral antidiabetic drugs: clinical relevance. Clin Pharmacokinet 44:1209–1225
Knapp RG, Miller MC (1992) Describing the performance of a diagnostic test. In: Knapp RG, Miller MC (eds) Clinical epidemiology and biostatistics, 1st edn. Williams & Wilkins, Baltimore, pp 41–42
Miners JO, Birkett DJ (1998) Cytochrome P4502C9: an enzyme of major importance in human drug metabolism. Br J Clin Pharmacol 45:525–538
Nathan DM, Buse JB, Davidson MB, Ferrannini E, Holman RR, Sherwin R et al (2009) Medical management of hyperglycaemia in type 2 diabetes mellitus: a consensus algorithm for the initiation and adjustment of therapy: a consensus statement from the American Diabetes Association and the European Association for the Study of Diabetes. Diabetologia 52:17–30
Nelson DR (2002) Mining databases for cytochrome P450 genes. Methods Enzymol 357:3–15
Niemi M, Cascorbi I, Timm R, Kroemer HK, Neuvonen PJ, Kivisto KT (2002) Glyburide and glimepiride pharmacokinetics in subjects with different CYP2C9 genotypes. Clin Pharmacol Ther 72:326–332
Robert F, Fendri S, Hary L, Lacroix C, Andréjak M, Lalau JD (2003) Kinetics of plasma and erythrocyte metformin after acute administration in healthy subjects. Diabetes Metab 29(3):279–283
Sullivan-Klose TH, Ghanayem BI, Bell DA, Zhang ZY, Kaminsky LS, Shenfield GM, Miners JO, Birkett DJ, Goldstein JA (1996) The role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics 6(4):341–349
Surendiran A, Pradhan SC, Agrawal A, Subrahmanyam DKS, Rajan S, Anichavezhi D, Adithan C (2011) Influence of CYP2C9 gene polymorphisms on response to glibenclamide in type 2 diabetes mellitus patients. Eur J Clin Pharmacol 67:797–801
Swen JJ, Wessels JA, Krabben A, Assendelft WJ, Guchelaar HJ (2010) Effect of CYP2C9 polymorphisms on prescribed dose and time-to-stable dose of sulfonylureas in primary care patients with type 2 diabetes mellitus. Pharmacogenomics 11(11):1517–1523
Toriello M, Meccariello P, Mazzaccara C, Di Fiore R, Esposito C, Sacchetti L (2006) Comparison of the TaqMan and LightCycler systems in pharmacogenetic testing: evaluation of CYP2C9*2/*3 polymorphisms. Clin Chem Lab Med 44(3):285–287
Trinder P (1969) Determination of blood glucose using an oxidaseperoxidase system with a non-carcinogenic chromogen. J Clin Path 22(2):158–161
Weise A, Prause S, Eidens M, Weber MM, Kann PH, Forst T, Pfützner A (2010) Prevalence of CYP450 gene variations in patients with type 2 diabetes. Clin Lab 56(7–8):311–318
World Medical Association (2008) Declaration of Helsinki. Ethical principles for medical research involving human subjects, the 59th WMA General Assembly, Seoul, South Korea
Yin O, Tomlinson B, Chow S (2005) CYP2C9, but not CYP2C19, polymorphisms affect the pharmacokinetics and pharmacodynamics of glyburide in Chinese subjects. Clin Pharmacol Ther 78:370–377
Zhou K, Donnelly L, Burch L et al (2009) Loss-of function CYP2C9 variants improve therapeutic response to sulfonylureas in type 2 diabetes (2009): a Go-DARTS Study. Clin Pharmacol Ther 87(1):52–56
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Salam, R.F.A., Zeyada, R. & Osman, N.A. Effect of CYP2C9 gene polymorphisms on response to treatment with sulfonylureas in a cohort of Egyptian type 2 diabetes mellitus patients. Comp Clin Pathol 23, 341–346 (2014). https://doi.org/10.1007/s00580-012-1620-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00580-012-1620-5