Abstract
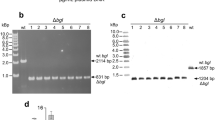
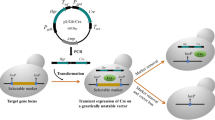
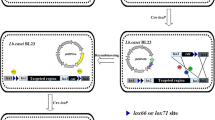
Markerless gene-disruption technology is particularly useful for effective genetic analyses of Thermus thermophilus (T. thermophilus), which have a limited number of selectable markers. In an attempt to develop a novel system for the markerless disruption of genes in T. thermophilus, we applied a Cre/lox system to construct a triple gene disruptant. To achieve this, we constructed two genetic tools, a loxP–htk–loxP cassette and cre-expressing plasmid, pSH-Cre, for gene disruption and removal of the selectable marker by Cre-mediated recombination. We found that the Cre/lox system was compatible with the proliferation of the T. thermophilus HB27 strain at the lowest growth temperature (50 °C), and thus succeeded in establishing a triple gene disruptant, the (∆TTC1454::loxP, ∆TTC1535KpnI::loxP, ∆TTC1576::loxP) strain, without leaving behind a selectable marker. During the process of the sequential disruption of multiple genes, we observed the undesired deletion and inversion of the chromosomal region between multiple loxP sites that were induced by Cre-mediated recombination. Therefore, we examined the effects of a lox66–htk–lox71 cassette by exploiting the mutant lox sites, lox66 and lox71, instead of native loxP sites. We successfully constructed a (∆TTC1535::lox72, ∆TTC1537::lox72) double gene disruptant without inducing the undesired deletion of the 0.7-kbp region between the two directly oriented lox72 sites created by the Cre-mediated recombination of the lox66–htk–lox71 cassette. This is the first demonstration of a Cre/lox system being applicable to extreme thermophiles in a genetic manipulation. Our results indicate that this system is a powerful tool for multiple markerless gene disruption in T. thermophilus.






Similar content being viewed by others
References
Albert H, Dale EC, Lee E, Ow DW (1995) Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J 7:649–659
Angelov A, Li H, Geissler A, Leis B, Liebl W (2013) Toxicity of indoxyl derivative accumulation in bacteria and its use as a new counterselection principle. Syst Appl Microbiol 36:585–592
Aoki K, Itoh T (2007) Characterization of the ColE2-like replicon of plasmid pTT8 from Thermus thermophilus. Biochem Biophys Res Commun 353:1028–1033
Averhoff B (2006) Genetic systems for Thermus. Methods Microbiol 35:279–308
Blas-Galindo E, Cava F, López-Viñas E, Mendieta J, Berenguer J (2007) Use of a dominant rpsL allele conferring streptomycin dependence for positive and negative selection in Thermus thermophilus. Appl Environ Microbiol 73:5138–5145
Brouns SJJ, Wu H, Akerboom J, Turnbull AP, de Vos WM, van der Oost J (2005) Engineering a selectable marker for hyperthermophiles. J Biol Chem 280:11422–11431
Buchholz F, Ringrose L, Angrand PO, Rossi F, Stewart AF (1996) Different thermostabilities of FLP and Cre recombinases: implications for applied site-specific recombination. Nucleic Acids Res 24:4256–4262
Buchholz F, Angrand PO, Stewart AF (1998) Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat Biotechnol 16:657–662
Carr JF, Danziger ME, Huang AL, Dahlberg AE, Gregory ST (2015) Engineering the genome of Thermus thermophilus using a counterselectable marker. J Bacteriol 197:1135–1144
Coppoolse ER, de Vroomen MJ, Roelofs D, Smit J, van Gennip F, Hersmus BJM, Nijkamp HJJ, van Haaren MJJ (2003) Cre recombinase expression can result in phenotypic aberrations in plants. Plant Mol Biol 51:263–279
Faraldo MM, de Pedro MA, Berenguer J (1992) Sequence of the S-layer gene of Thermus thermophilus HB8 and functionality of its promoter in Escherichia coli. J Bacteriol 174:7458–7462
Fujita A, Sato T, Koyama Y, Misumi Y (2015) A reporter gene system for the precise measurement of promoter activity in Thermus thermophilus HB27. Extremophiles 19:1193–1201
Henne A, Brüggemann H, Raasch C, Wiezer A, Hartsch T, Liesegang H, Johann A, Lienard T, Gohl O, Martinez-Arias R, Jacobi C, Starkuviene V, Schlenczeck S, Dencker S, Huber R, Klenk HP, Kramer W, Merkl R, Gottschalk G, Fritz HJ (2004) The genome sequence of the extreme thermophile Thermus thermophilus. Nat Biotechnol 22:547–553
Hiratsu K, Shiotani S, Makino K, Nunoshiba T (2013) Construction of a supF-based system for detection of mutations in the chromosomal DNA of Arabidopsis. Mol Genet Genom 288:707–715
Hoess RH, Abremski K (1985) Mechanism of strand cleavage and exchange in the Cre-lox site-specific recombination system. J Mol Biol 181:351–362
Hoseki J, Yano T, Koyama Y, Kuramitsu S, Kagamiyama H (1999) Directed evolution of thermostable kanamycin-resistance gene: a convenient selection marker for Thermus thermophilus. J Biochem 126:951–956
Kovács ÁT, van Hartskamp M, Kuipers OP, van Kranenburg R (2010) Genetic tool development for a new host for biotechnology, the thermotolerant bacterium Bacillus coagulans. Appl Environ Microbiol 76:4085–4088
Koyama Y, Hoshino T, Tomizuka N, Furukawa K (1986) Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J Bacteriol 166:338–340
Lambert JM, Bongers RS, Kleerebezem M (2007) Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl Environ Microbiol 73:1126–1135
Leibig M, Krismer B, Kolb M, Friede A, Götz F, Bertram R (2008) Marker removal in staphylococci via Cre recombinase and different lox sites. Appl Environ Microbiol 74:1316–1323
Leis B, Angelov A, Li H, Liebl W (2014) Genetic analysis of lipolytic activities in Thermus thermophilus HB27. J Biotechnol 191:150–157
Nagy A (2000) Cre recombinase: the universal reagent for genome tailoring. Genesis 26:99–109
Nakamura A, Takakura Y, Kobayashi H, Hoshino T (2005) In vivo directed evolution for thermostabilization of Escherichia coli hygromycin B phosphotransferase and the use of the gene as a selection marker in the host-vector system of Thermus thermophilus. J Biosci Bioeng 100:158–163
Nakane S, Nakagawa N, Kuramitsu S, Masui R (2012) The role of the PHP domain associated with DNA polymerase X from Thermus thermophilus HB8 in base excision repair. DNA Repair (Amst) 11:906–914
Ohta T, Tokishita S, Imazuka R, Mori I, Okamura J, Yamagata H (2006) β-Glucosidase as a reporter for the gene expression studies in Thermus thermophilus and constitutive expression of DNA repair genes. Mutagenesis 21:255–260
Ohtani N, Tomita M, Itaya M (2010) An extreme thermophile, Thermus thermophilus, is a polyploid bacterium. J Bacteriol 192:5499–5505
Oshima T, Imahori K (1974) Description of Thermus thermophilus (Yoshida and Oshima) comb. nov., a nonsporulating thermophilic bacterium from a Japanese thermal spa. Int J Syst Bacteriol 24:102–112
Sakai T, Tokishita S, Mochizuki K, Motomiya A, Yamagata H, Ohta T (2008) Mutagenesis of uracil-DNA glycosylase deficient mutants of the extremely thermophilic eubacterium Thermus thermophilus. DNA Repair (Amst) 7:663–669
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Schmidt EE, Taylor DS, Prigge JR, Barnett S, Capecchi MR (2000) Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc Natl Acad Sci USA 97:13702–13707
Short JM, Fernandez JM, Sorge JA, Huse WD (1988) λ ZAP: a bacteriophage λ expression vector with in vivo excision properties. Nucleic Acids Res 16:7583–7600
Sternberg N, Hamilton D (1981) Bacteriophage P1 site-specific recombination: I. Recombination between loxP sites. J Mol Biol 150:467–486
Suzuki N, Nonaka H, Tsuge Y, Inui M, Yukawa H (2005) New multiple-deletion method for the Corynebacterium glutamicum genome, using a mutant lox sequence. Appl Environ Microbiol 71:8472–8480
Tamakoshi M, Yaoi T, Oshima T, Yamagishi A (1999) An efficient gene replacement and deletion system for an extreme thermophile, Thermus thermophilus. FEMS Microbiol Lett 173:431–437
Wang L, Hoffmann J, Watzlawick H, Altenbuchner J (2016) Markerless gene deletion with cytosine deaminase in Thermus thermophilus strain HB27. Appl Environ Microbiol 82:1249–1255
Yan X, Yu HJ, Hong Q, Li SP (2008) Cre/lox system and PCR-based genome engineering in Bacillus subtilis. Appl Environ Microbiol 74:5556–5562
Yanisch-Perron C, Vieira J, Messing J (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119
Yokoyama S, Hirota H, Kigawa T, Yabuki T, Shirouzu M, Terada T, Ito Y, Matsuo Y, Kuroda Y, Nishimura Y, Kyogoku Y, Miki K, Masui R, Kuramitsu S (2000) Structural genomics projects in Japan. Nat Struct Mol Biol 7:943–945
Acknowledgements
We thank Prof. Akira Nakamura (University of Tsukuba) for kindly providing the pT8H5-Pslp plasmid. We also thank Prof. Toshihiro Ohta (Tokyo University of Pharmacy and Life Science) for kindly providing the pTAP60 plasmid and wild-type T. thermophilus HB27 strain.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Togawa, Y., Nunoshiba, T. & Hiratsu, K. Cre/lox-based multiple markerless gene disruption in the genome of the extreme thermophile Thermus thermophilus . Mol Genet Genomics 293, 277–291 (2018). https://doi.org/10.1007/s00438-017-1361-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-017-1361-x