Abstract
In this study, we explore blood parasite prevalence, infection intensity, and co-infection levels in an urban population of feral pigeons Columba livia in Cape Town. We analyze the effect of blood parasites on host body condition and the association between melanin expression in the host’s plumage and parasite infection intensity and co-infection levels. Relating to the haemosporidian parasite itself, we study their genetic diversity by means of DNA barcoding (cytochrome b) and show the geographic and host distribution of related parasite lineages in pigeons worldwide. Blood from 195 C. livia individuals was collected from April to June 2018. Morphometric measurements and plumage melanism were recorded from every captured bird. Haemosporidian prevalence and infection intensity were determined by screening blood smears and parasite lineages by DNA sequencing. Prevalence of Haemoproteus spp. was high at 96.9%. The body condition of the hosts was negatively associated with infection intensity. However, infection intensity was unrelated to plumage melanism. The cytochrome b sequences revealed the presence of four Haemoproteus lineages in our population of pigeons, which show high levels of co-occurrence within individual birds. Three lineages (HAECOL1, COLIV03, COQUI05) belong to Haemoproteus columbae and differ only by 0.1% to 0.8% in the cytochrome b gene. Another lineage (COLIV06) differs by 8.3% from the latter ones and is not linked to a morphospecies, yet. No parasites of the genera Leucocytozoon and Plasmodium were detected.
Similar content being viewed by others
Introduction
Wild feral pigeon populations of Columba livia f. domestica Gmelin, 1789 are typical inhabitants of urban landscapes, where they are associated with humans due to the availability of suitable nesting sites and anthropogenic food. These factors have enabled feral pigeons and many of its parasites to colonize cities across the globe (Johnston and Janiga 1995). The undomesticated relative is the rock pigeon Columba livia, whose native range is restricted to Western and Southern Europe. Archeological evidence suggests that rock pigeons were domesticated several thousand years ago (Shapiro and Domyan 2013) and have been used as a food source as early as 10,000 years ago (Blasco et al. 2014). Since then, feral pigeons have successfully expanded their range and established wild populations worldwide, mainly in urban environments. The feral pigeon is not native to South Africa but was introduced as a domestic species in the seventeenth century (Brooke 1981; Dean 2000). Cape Town is a hotspot of biodiversity (Myers et al. 2000) and of special interest as an urban study location. Not only is the literature body comparatively scarce in the “Global South” in comparison to the “Global North” (Hedblom and Murgui 2017), but urban environments are also rapidly changing in the Southern Hemisphere (Swilling 2006). Feral pigeons are likely the most successful avian urban exploiters that manage very well in urban and suburban environments. Furthermore, they are also a model organism in many research disciplines, i.e., parasitology, behavioral science, and physiology (i.e., Abs 1983; Harbison et al. 2009; Klopfleisch et al. 2006).
Avian haemosporidians are comprised of apicomplexan protozoans of the genera Plasmodium, Haemoproteus, and Leucocytozoon. Haemosporidians are the most diverse group of avian blood parasites with several hundred species infecting birds all over the world (Valkiūnas 2004). The genus Haemoproteus contains two subgenera: Haemoproteus and Parahaemoproteus (Valkiūnas 2004). Haemoproteus comprises about ten parasite species described from Columbidae hosts and one species each from Laridae and Fregatidae hosts. Parahaemoproteus comprises the majority of Haemoproteus parasite specialized on passerines (Levin et al. 2012; Levin et al. 2011; Valkiūnas 2004; Valkiūnas et al. 2013; Valkiūnas et al. 2010). The two Haemoproteus subgenera form distinct clades in molecular phylogenetic analyses (Galen et al. 2018; Pacheco et al. 2017) and depend on different dipteran groups as vectors. Species of the subgenus Haemoproteus are transmitted by dipterans of the family Hippoboscidae, whereas species of the subgenus Parahaemoproteus are transmitted by biting midges of the family Ceratopogonidae (genus Culicoides).
Haemoproteus columbae Kruse, 1890 was described from C. livia and is transmitted horizontally by blood-sucking insects, predominantly by the ectoparasitic pigeon louse fly Pseudolynchia canariensis Macquart, 1839 (Sol et al. 2000). Blood stages of Haemoproteus species are quite conspicuous in blood smears (Valkiūnas 2004), but species determination is complicated due to the limited number of distinct morphological features. More recently, DNA sequencing using PCR has been established to identify blood parasite lineages (Fallon et al. 2003) and has also been used as a more accurate method for identifying blood parasite prevalence, especially at low infection intensities (Fallon and Ricklefs 2008; Fallon et al. 2003; Garamszegi 2010). Furthermore, genetic sequencing enables the identification of closely related blood parasite lineages, which cannot be differentiated through microscopic examination. Sequencing also allows the determination of co-infections of multiple haemosporidian lineages, which commonly occur (Alizon et al. 2013; Poulin and Morand 2000; Silva-Iturriza et al. 2012). To distinguish avian haemosporidian species or lineages, the mitochondrial cytochrome b (cytb) is often used as a genetic marker as it incorporates the so-called “DNA barcode” region for haemosporidians (Bensch et al. 2009; Dimitrov et al. 2014). Single and multiple infections of several blood parasite lineages can have different effects on the hosts and have been associated with negative effects on their body conditions (Marzal et al. 2008), lower hematocrit values (Palinauskas et al. 2011), and lower host survival (Pigeault et al. 2018). However, in studies on house martins Delichon urbica, birds with double infections showed better reproductive performance (Marzal et al. 2008; Pigeault et al. 2018).
Generally, H. columbae infections are not considered to be very detrimental to their hosts (Bennett et al. 1993; Earle et al. 1993; Sol et al. 2003). However, high infection intensity can be associated with negative physiological (Earle et al. 1993) and behavioral (Markus and Oosthuizen 1972) changes. In some cases, infections can even be lethal, especially in young individuals that have not yet developed an adequate immune response (Sol et al. 2003). Due to their successful expansion across the globe, feral pigeons act as vectors for H. columbae and other haemosporidian parasites. They can facilitate the spread into naïve new hosts and can potentially threaten local columbid bird species (Chagas et al. 2016; Foronda et al. 2004; Lee-Cruz et al. 2016; Peirce et al. 1997; Swinnerton et al. 2005; van Riper et al. 1986). Specifically in Cape Town, feral pigeons regularly form mixed flocks with the native speckled pigeon Columba guinea and might allow the spread of feral pigeon–specific blood parasites to naïve hosts (Earle and Little 1993; Little 1994). In an earlier study, Haemoproteus has been identified in speckled pigeons; however, this study was based on blood smear analysis only. In the absence of DNA sequencing, it remains unknown whether these represent blood parasite lineages originating from feral pigeons or whether they are unique to native speckled pigeons. Furthermore, feral pigeons were identified as potential vectors of novel parasites for endemic and endangered columbid species native in the Canary Islands (Foronda et al. 2004). Haemoproteus columbae has not yet been found in a non-columbid host and might be specific to pigeon and dove hosts (Valkiūnas 2004).
The domesticated feral pigeon is color polymorphic, whereby different morphs are independent of age or sex, are interbreeding freely, and can be present within the same population (Huxley 1955). The species features a wide range of plumage colorations ranging from completely white to almost fully melanistic (Haase et al. 1992; Jacquin et al. 2011, 2013; Johnston and Janiga 1995). Plumage phenotypes are determined by eumelanin and pheomelanin that are known for their pleiotropic effects on the immune system through the melanocortin system (Ducrest et al. 2008), whereby the gene coding the phenotype also controls the expression of several different traits, unrelated to color, such as immunocompetence. Plumage variation in the feral pigeon is associated with a stronger immune system (Jacquin et al. 2011), and white pigeons (i.e., lacking melanin) were found to have a higher blood parasite prevalence with increasing levels of urbanization (Jacquin et al. 2013). In turn, darker individuals showed a stronger cortisone stress response in rural areas than in urban habitats (Corbel et al. 2016).
In the present study, (1) we describe H. columbae prevalence and infection intensity in a wild feral pigeon population in the city of Cape Town, South Africa. Additionally, (2) we assess blood parasite diversity by PCR screening for H. columbae lineages and by sequencing a section of the mitochondrial cytb gene. (3) We show the geographic and host distribution of H. columbae and related parasite lineages in pigeons worldwide. (4) We test for potential health consequences of blood parasite infection by exploring whether higher infection intensity and co-infection with different Haemoproteus lineages reduce individual body condition. We hypothesize that those blood parasites have a negative effect on the health of the host and thus predict that individual body condition decreases with increasing infection intensity or infection by multiple parasite species. Lastly, (5) we explore for potential pleiotropic effects of color polymorphism by correlating infection intensity and co-infection with the expression of eumelanin. Here, we hypothesize that eumelanin expression influences an individual’s health. Therefore, we predict that feral pigeons with darker plumage coloration have lower haemosporidian infection intensity and a lower number of co-infections.
Material and methods
Study site and feral pigeons
For this study, 195 wild feral pigeons were caught in walk-in traps in suburban areas of Cape Town, South Africa (33.92° S, 18.42° E) from April to June 2018, during the austral autumn and winter. We attempted to obtain data for all birds on morphometric measurements, blood samples for smears, and DNA extraction; however, some data is missing from some birds due to time constraints in processing several individuals in a short period of time. Sample sizes (n) are given in brackets below throughout; for a complete sample list, see Appendix Table S1.
Before blood sampling, pigeons were routinely housed communally in a pigeon coop at Eagle Encounters, Stellenbosch (33.97° S, 18.78° E), for a maximum of 6 days prior to being part of an unrelated behavioral experiment (Nebel et al. 2019). A maximum of 1 ml blood was taken from the brachial vein of each individual, and these blood samples were then immediately use to prepare blood smears on microscope slides and air-dried. Remaining blood was stored in EDTA buffer at 4 °C. We measured the tarsus length with a caliper (to the closest 0.1 mm) and weighed the pigeon on a scale (in g). These measures were taken to derive individual body condition and were obtained from 187 individuals in total. The age of the pigeons was classified depending on the color of the eyes and cere (immature, < 6 months of age, n = 12, or adult, > 6 months, n = 183; following Kautz and Seamans 1986). Sex was not determined. All pigeons were individually marked and released back at the source of capture.
Assessment of plumage phenotype
Plumage coloration in the feral pigeon is determined by eumelanin and pheomelanin deposited in the feathers (Haase et al. 1992). Although both eumelanin and pheomelanin are associated with the melanocortin system, and both can have beneficial effects on the immune system (Ducrest et al. 2008; Jacquin et al. 2011), in this study, we focus on pigeons showing a variation of eumelanin and excluded all individuals with a pheomelanin-based plumage (i.e., reddish plumage; n = 3) due to the small sample size. Systematic photographs were taken (showing the belly, back with one wing spread and the head) to document the plumage of every individual. The feral pigeons’ phenotypes were determined in a twofold approach. First, melanin content of the wing was quantified by using the software ImageJ (Abramoff et al. 2004) and then categorized according to levels of melanism by author AP following established protocols of Johnston and Janiga (1995) and Jacquin et al. (2011): phenotypes 0 (“white,” n = 1), 1 (“blue bar,” n = 40), 2 (“checker,” < 50% dark feathers on the wing surface, n = 57), 3 (“T-pattern,” > 50% dark feathers on the wing surface, n = 65), and 4 (completely melanistic, n = 28). The plumage of one pigeon could not be scored, because no photos were taken during processing. One individual showing mainly white feathers (score 0) was excluded from all analyses due to low sample size. Ten percent of all individuals were randomly re-scored, and the assignment was highly reproducible.
Assessment of infection intensity by blood slide screening
Blood smears of 192 individual C. livia were fixed with methanol and stained with Giemsa stain following the standard protocol of Hemacolor® Rapid staining of blood smear kit (Merck, Darmstadt, Germany). The slides were first scanned under the microscope at × 400 magnification to determine the presence or absence of haemosporidian blood parasites. For infected individuals, the intensity of infection was then determined by scanning each slide at × 1000 magnification with an oil immersion lens and counting the number of parasites seen within 10,000 erythrocytes.
Molecular genetics: assessment parasite lineages and co-infections
Molecular screening for blood parasites and identification of Haemoproteus lineages were performed on 144 feral pigeon samples of which aliquots of blood were stored in EDTA buffer (in 51 cases, all obtained blood was used up, preparing the blood smears). DNA was extracted with the DNeasy Blood & Tissue Kit (Qiagen, Venlo, Netherlands) following the standard protocol for isolation from blood and using equivalents of about 10 μl blood from each sample. We amplified an 886-bp section of the mitochondrial cytb using the primers CytB_HPL_intF1 (5′-GAGAATTATGGAGTGGATGGTG-3′) and CytB_HPL_intR1 (5′-ATGTTTGCTTGGGAGCTGTAATC-3′) (Harl et al. 2019), which covers the so-called DNA barcode region for avian haemosporidians. CytB_HPL_intF1 binds at nucleotide positions 174 to 195 of the cytb, and CytB_HPL_intR1 binds at a conserved site 13-bp from the 3′ end of the cytb. PCRs were performed with the GoTaq Long PCR Master Mix (Promega, Madison, Wisconsin, USA) on a peqSTAR 2X Universal Gradient thermocycler (VWR, Radnor, Pennsylvania, USA). Each 25 μl master mix contained 12.5 μl GoTaq Long PCR Master Mix, 9.5 μl H2O, each 1 μl of 10 mM primer, and 1 μl of DNA template. The PCRs started with an initial denaturation for 2 min at 94 °C, followed by 35 cycles with 30 s at 94 °C, 30 s at 53 °C, 1 min at 65 °C, and a final extension for 10 min at 72 °C. PCR products were visualized on 1% agarose gels, and positive samples were purified with the QIAquick PCR Purification Kit (Qiagen). Sequencing was performed at Microsynth Austria GmbH (Vienna, Austria) using the PCR primer CytB_HPL_intR1. Raw sequences were inspected by eye with BioEdit v.7.0.8 (Hall 1999). Sequences of samples with double and triple infections featured ambiguous sites with wobble bases, which were consequently checked in the electropherograms. After the first inspection, the sequences were sorted by similarity and all aberrant sites were rechecked. Identification (and unphasing) of parasite lineages in samples with multiple infections was straightforward, because the number of different haplotypes in our sample was low. Haplotype networks, based on a 478-bp cytb fragment, were calculated with two different datasets, the first comprising haplotypes related to H. columbae and the second comprising the lineage of a yet unidentified Haemoproteus species (COLIV06) and related haplotypes. To visualize the geographic and host distribution, we included already published sequences from GenBank (NCBI) in the networks. These were retrieved by performing BLAST searches and including all samples with sequence similarity of 97% to 100% to haplotypes found in the present study. Only those BLAST hits were kept, which did not contain ambiguities and which covered the 476-bp DNA barcode region of the cytb. The respective lineage names of the haplotypes were retrieved by BLAST searches against the MalAvi database (Bensch et al. 2009). The sequences were aligned with MAFFT v.7 (Katoh and Standley 2013) and trimmed to the size of the barcode region. Median-Joining networks were calculated with Network v.5.1.0.0 (Fluxus Technology Ltd., Suffolk, England) applying the default settings. In order to reduce unnecessary median vectors, the networks were post-processed with the maximum parsimony (MP) option. Networks were graphically prepared and supplemented by information on host species and geographic region in Network Publisher v.2.1.1.2 (Fluxus Technology Ltd.) and finalized in Adobe Illustrator CC v.19.0.0 (San Jose, California, USA). Co-infection was determined based on DNA sequencing and reflects the number of Haemoproteus lineages (1 = mono-infection, 2 = double-infection, and 3 = triple-infection).
Statistical analysis
We explored whether individual body condition was associated with blood parasite parameters (i.e., infection intensity or co-infection level, models a and b) using linear models (LMs), and whether blood parasite parameters were associated with plumage phenotype using LM (model c) and a multinomial logistic regression (model d). Linear models were fit with the stats package (version 3.4.0) and the multinomial regression nnet package (7.3-12) (Ripley et al. 2016) in the R environment, version 3.4.4 (R Core Team 2018). All quantitative variables (body mass, tarsus length, and infection intensity) were scaled beforehand. Six individuals that showed no prevalence in the blood smear analysis (even if they were positives in the PCR screening) were excluded from all models featuring infection intensity. However, four of these were included in all models exploring the effects of co-infection on body condition and the effect of melanism expression on co-infection, because sequence data were available.
Previous research has shown that juvenile individuals are often suffering from higher infection intensities and are in lower body condition than adults (Sol et al. 2003). Therefore, before running the main analyses, we explored the influence of age-specific differences on individual body condition and infection intensity. Two linear models were fitted: one with body condition and a second with infection intensity (log-transformed to ensure normally distributed residuals) as response variables. Age was the key explanatory variable (immature or adult), and the length of the tarsus was added as a control variable in the model exploring body condition to control for individual variation in size. Due to a significant difference between immature and adult pigeons (see the “Results” section) and small sample size, immature individuals (n = 12) were excluded from the main models introduced below.
In the first set of the main analysis, we explored the effect of infection intensity (model a, n = 169) and co-infection (model b, n = 125) on individual condition and we fitted a linear model with body mass as the response variable and infection intensity (%) or co-infection of Haemoproteus spp. lineages (categorical variable, mono-, double-, or triple-infection) as explanatory variables. The length of the tarsus and the phenotype were also added as covariates to control for the individual size of the pigeon and to take the effect of different melanin expression levels into account. This approach was generally favored, rather than fitting residuals of a regression between body mass and tarsus as a body condition response in the model (García-Berthou 2001; Smith 1999).
In a second set of analysis, we explored the effect of plumage phenotype on blood parasite parameters, infection intensity (model c, n = 169), and co-infection (model d, n = 130). To do so, we fitted a linear model with log-transformed infection intensity as a response in order to fulfill the requirement of normally distributed residuals. In all linear models, residuals were visually inspected to ensure a normal distribution. Additionally, we explored whether pigeons of different phenotypes were infected by different numbers of blood parasites by fitting a multinomial logistic regression with the number of lineages as the response variable. Infection with only one parasite was the reference. p values were obtained by performing a Wald test (z test). In both models, the degree of melanism (phenotypes 1–4) was fitted as a continuous explanatory variable. As infection intensity might be influenced by multiple competing parasites (Clark et al. 2016), we fitted the co-infection variable as a fixed effect in explorative models a and c, and co-infection did not improve the model fit (by AIC) and was returned as not significant. It was thus removed from the final models.
Results
Haemosporidian parasite infection intensity and prevalence
Blood smears were prepared for 192 feral pigeons and scored for haemosporidian parasite infections. In the blood smear analysis, only six individuals, out of 192, were identified as uninfected (no parasites detected in 10,000 erythrocytes, 96.9% infection prevalence). However, four of these six birds did score positive in the PCR screening. Molecular screening could not be performed for one sample due to insufficient blood volume, and for a second one, the repeated PCR was negative. All positive samples were infected with Haemoproteus spp., whereas Leucocytozoon spp. and Plasmodium spp. were absent in our study population. The mean infection intensity of Haemoproteus spp. positive individuals was 1.3% (SD 2.9), ranging from 0.007 and 20.4% of erythrocytes being infected. The distribution of infection intensity was skewed, and the median was lower than the mean (median = 0.3%).
Molecular analysis of haemosporidian parasites
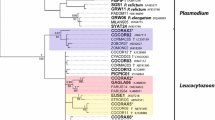
Molecular screening for blood parasite lineages was performed on 144 DNA samples isolated from pigeon blood. PCRs for all but one sample were positive, and products were sequenced from 143 samples. Almost all samples contained lineages belonging to H. columbae (HAECOL1, COLIV03, COQUI05), which is a common parasite of pigeons, and only four samples contained another Haemoproteus lineage (COLIV06) not yet linked to a morphospecies. A high number of pigeons were infected with more than one parasite lineage. From the 143 PCR positive samples, 81 (56.6%) featured a single lineage, 51 (35.7%) two lineages, and 11 (15.7%) three lineages. The most common lineage HAECOL1 was found in 133 pigeons. We actually found two haplotypes matching 100% with HAECOL1, but differing in a single site at position 1003 of the cytb. The first variant (HAECOL1a) was found in 110 specimens and the second one (HAECOL1b) in 37 birds, and both haplotypes co-occurred in 16 individual pigeons. In the following, we refer to both of the latter haplotypes as lineage HAECOL1. The MalAvi lineages COLIV03 and COQUI05 were found in 44 and 36 samples, respectively. At the 478-bp DNA barcode section, HAECOL1 and COQUI05 differ from each other in three sites (0.6% uncorrected p distance), HAECOL1 and COLIV03 in four sites (0.8%), and COLIV03 and COQUI05 in five sites (1%). Only four pigeons were infected with the lineage COLIV06 belonging to an unidentified Haemoproteus species, differing from the H. columbae lineages by about 10%. The relationship of haplotypes found in the present study and already published data is displayed in Median-Joining networks (Fig. 1). All cytb sequences are available at NCBI GenBank under the accession numbers MN065190–MN065419.
Median-joining haplotype networks of cytb sequences (476-bp) from the present study and GenBank. The size of the circles corresponds to the number of identical haplotypes. Bars on branches connecting the circles indicate the number of differences (substitutions) between haplotypes. Small white circles indicate median vectors. If available, the MalAvi lineage names are provided for each haplotype. Networks on the left side show haplotypes of H. columbae and related lineages, and networks on the right side show haplotypes of Haemoproteus spp. (COLIV06) and related lineages. a Information on host species. b Geographic distribution
Relationship between parasite infection, plumage phenotype, age, and body condition
Young pigeons (n = 12) had a significantly lower body condition (immature, 292.5 (SD 51.4); adult, 332.1 (SD 43.7); χ2 = 6.9, estimate = − 0.8 (SE 0.3), n = 174, p = 0.002) and higher infection intensity (immature, 5.7% (SD 5.7); adult, 1.0% (SD 2.4); χ2 = 53.2, estimate = 2.2 (SE 0.5), n = 174, p < 0.001). These young birds were excluded from the subsequent analysis. There was a significant (linear) negative relationship between infection intensity and body condition. Body mass decreased by 5.24 g with a 1% increase of infected red blood cell (Table 1, Fig. 2).
We found no relationship between the levels of co-infection of Haemoproteus spp. (mono-, double-, or triple-infection) and body condition (Table 1). We also found no relationship between plumage phenotype and infection intensity or co-infection levels (Table 1). Feral pigeons with a paler plumage had similar levels of infection intensity and co-infection as conspecifics with darker melanic plumage (Table 2).
Discussion
Based on the blood smear analysis, the prevalence of Haemoproteus spp. was high in our study population with a 96.9% infection rate. Only six individuals did not show any infection with haemosporidian parasites based on the blood smear screening, but four of these tested positive through molecular screening. Screenings of blood smears often underestimate infection prevalence—especially when infection intensity is low—whereas PCR is more sensitive (Valkiūnas et al. 2008). Our results thus show that almost all feral pigeons sampled in suburban Cape Town were infected with H. columbae. This high rate is in line with results of a study conducted in Pretoria in the 1970s with 100% (Markus and Oosthuizen 1972), but much higher than previously reported in Cape Town in the 1990s with 72.7% infection rate (Earle and Little 1993). Both studies used only blood smear screenings and did not apply any PCR-based methods. Similarly high prevalence values were found in cities outside of South Africa, i.e., 97–100% in Madrid, Barcelona, and Granollers (Spain) (Sol et al. 2000; Vázquez et al. 2010); 100% in Sao Paulo (Brazil) (Chagas et al. 2016); or 100% in Hyattsville (Maryland, USA) (Knisley and Herman 1967). Haemoproteus spp. infection intensities of C. livia have been studied in other Sub-Saharan African countries with prevalence levels varying greatly both between and within countries. Haemoproteus spp. prevalence was high at 75–80% in Sebele (Botswana), Owerri (Nigeria), and Kampala (Uganda) (Dranzoa et al. 1999; Mushi et al. 2000; Opara et al. 2012); intermediate at 50% in Kano (Nigeria) (Karamba et al. 2012), and 37% in Morogoro (Tanzania) (Msoffe et al. 2010); and low at 15.2% in Zaria (Nigeria) (Owolabi et al. 2009). High prevalence levels and infection intensities of Haemoproteus spp. are not surprising, given that pigeons live and roost in flocks, making parasite transmission very easy (Johnston and Janiga 1995). Possibly, the high infection prevalence is depending on population density. Both Haemoproteus spp. prevalence and feral pigeon population density are generally high in suburban and urban studies (Chagas et al. 2016; Dranzoa et al. 1999; Karamba et al. 2012; Knisley and Herman 1967; Mushi et al. 2000; Opara et al. 2012; Sacchi et al. 2002; Sol et al. 2000; Vázquez et al. 2010). It can be assumed that these populations of feral pigeons are more likely to become infected by parasites and diseases because of their larger population densities. However, since natural populations of C. livia are poorly investigated, there is no data to compare with. Haemoproteus columbae does not infect non-columbid birds and thus is not a threat to other bird groups. However, breeding pigeons or native, naïve pigeon species are prone to become infected as the close proximity between wildlife, livestock, and pets could facilitate cross-species disease transmission in suburban and urban environments (Earle and Little 1993).
Although not much sequence data on avian haemosporidians of pigeons has been published so far, the four cytb lineages found in the present study seem to be common in feral pigeons. Exactly the same lineages were isolated from feral pigeons in the Sao Paolo Zoo (Chagas et al. 2016). The authors of the latter study confirmed that the lineages HAECOL1, COQUI05, and COLIV03 morphologically belong to H. columbae. The similarity in parasite lineage composition should be viewed in a historical context, since feral pigeons were introduced into both South Africa and South America by humans (Johnston and Janiga 1995), most likely together with their louse fly (Hippoboscidae) vectors. Apart from the four H. columbae lineages, only two other haplotypes are so far published for the feral pigeon: COLIV07 from Peru (Pacheco et al. 2017), differing in one site from HAECOL01, and an unnamed haplotype isolated from a pigeon in Japan (LC325859; published on GenBank only), differing in a single site from COQUI05. Related haplotypes have been isolated from common wood pigeon Columba palumbus in Europe (Dunn et al. 2017), African olive pigeon Columba arquatrix in Malawi (Lutz et al. 2015), Amboyna cuckoo-dove Macropygia amboinensis in Papua New Guinea (Beadell et al. 2004), band-tailed pigeon Patagioenas fasciata in Peru (JQ988683, JQ988729; published on GenBank only), and two mosquitoes (Culicidae, Diptera) in Cameroon (Njabo et al. 2011). Since the actual vector of H. columbae is the pigeon louse fly P. canariensis (Brooke 1981; Dean 2000; Sol et al. 2000), the parasites were probably in the blood meal of the mosquitoes. Interestingly, seven other related lineages (differing in 2 to 23 sites from HAECOL1) were found in both domestic and wild forms of the chicken Gallus gallus in Malaysia (Gimba et al., data published on GenBank only: KT290923–KT290928), which were not considered as hosts of H. columbae so far. Other than H. columbae, the second Haemoproteus species (lineage COLIV06) was found only in Columbidae birds. COLIV06 was elsewhere found in feral pigeons from Brazil, and several related lineages (differing in eight to 14 sites) were isolated in C. palumbus and European turtle-dove Streptopelia turtur in England (Dunn et al. 2017) and Spain (Drovetski et al. 2014); in red-eyed dove Streptopelia semitorquata, C. arquatrix, and African green pigeon Treron calvus in Malawi (Lutz et al. 2015); and in island bronze-naped pigeon Columba malherbii in Sao Tome and Principe (KT376899; published on GenBank only).
In our study, young pigeons had a significantly lower body condition and higher infection intensity than older individuals, a pattern often reported in different bird species (Sol et al. 2003). In adults, individual body condition was negatively correlated with infection intensity of Haemoproteus—implying that there may well be a physiological cost associated with being heavily infected. Parasites might drain energy from their host (Price et al. 1986), or infection could directly impair foraging activities (Marzal et al. 2005) and thus negatively affect body condition. To our knowledge, no correlation between Haemoproteus spp. and body condition in feral pigeons has been reported to date. The influence of different Parahaemoproteus parasites and infection intensity in non-columbids on body condition is frequently reported; however, these associations are not consistent. Negative effects of infection intensity on body condition have been found in American kestrel Falco sparverius (females only) (Dawson and Bortolotti 2000), but not in Eurasian kestrel Falco tinnunculus (Korpimäki et al. 1995) nor for European blackcap Sylvia atricapilla (during the first days after infection with Haemoproteus belopolskyi) (Valkiūnas et al. 2006). Furthermore, no significant negative correlations between infection intensity and body condition were documented in several other passerine species (Bennett et al. 1988; Granthon and Williams 2017), including during migration (correlation with body fat scores) (Ashford 1971). In the present study, co-infections with several Haemoproteus lineages did not affect the body condition more than infections with a single lineage only, which is in line with findings of Palinauskas et al. (2011) and Marzal et al. (2008). For future work, it would be interesting to investigate whether closely related lineages interact with each other. This interaction could be positive and favor co-infections, or it could be negative and closely related parasite lineages could compete within the host or within the vector. An example of a positive interaction was found in experimentally infected passerines. Two Plasmodium spp. acted synergetically during primary infections and were found to be highly virulent (Palinauskas et al. 2011). In a cross-genus study, microfilariae parasites were enhancing the co-infection of certain, but not all haemosporidians (Clark et al. 2016). In the present study, sequences were obtained by standard PCR and Sanger sequencing. Mixed infections were identified by the presence of wobble bases in the electropherograms, but we did not quantify the amount of each lineage in samples with mixed infections. It should be mentioned that with standard PCR and Sanger sequencing, parasites at much higher parasitemia (quantity) may be amplified preferentially, masking the fact that other parasites are present. In order to overcome this problem in future studies, deep amplicon sequencing would provide a possibility to accurately determine the percentage of two or more pathogen lineages in samples with mixed infections (Olmstead et al. 2019).
The feral pigeons caught for this study are part of a wild population in an area with large populations of native avian predator species (e.g. black sparrowhawk Accipiter melanoleucus (Martin et al. 2014; Rose et al. 2017); peregrine falcon Falco peregrinus (Jenkins 2000)), which may explain the absence of individuals with extremely high infection intensities of 50% or higher, since such birds might appear physically “sick” and might be rapidly removed from the system (Earle et al. 1993; Markus and Oosthuizen 1972). Most individuals sampled are likely exhibiting chronic infections rather than primary infections that are usually characterized by higher infection intensities (Asghar et al. 2012). Individuals during primary infections that show extreme infection intensities are more likely to die due to predation (Møller and Nielsen 2007; Temple 1987) or through the effects of the parasite themselves, like anemia (Earle et al. 1993). Our results show that H. columbae infection intensity has negative effects on a body condition which could lower individual fitness (Chastel et al. 1995; Cichoiń et al. 1998). However, our study does not include any long-term effects on individual survival, mate choice, or reproductive success and no such studies have been conducted to date on the H. columbae-C. livia parasite-host system. However, long-term fitness correlations have been studied in other species, for example female Eurasian kestrels mating with males infected with Haemoproteus (Parahaemoproteus) tinnunculi laid later and smaller clutches than females mating with uninfected males (Korpimäki et al. 1995), thus suggesting that Haemoproteus spp. infections play a role in mate choice. Prevalence of Haemoproteus (Parahaemoproteus) payevskyi in female great reed warblers Acrocephalus arundinaceus led to a smaller number of fledged offspring (Asghar et al. 2011), and high infection intensity of Haemoproteus spp. negatively influenced the return rates of female American kestrels, implying that infection reduced survival rates (Dawson and Bortolotti 2000). On the contrary, white-crowned sparrow Zonotrichia leucophrys oriantha females infected with Haemoproteus (Parahaemoproteus) beckeri had higher survival rates and about twice as high lifetime reproductive success than uninfected females (Zylberberg et al. 2015).
Interestingly, other avian malaria parasites like Leucocytozoon spp. and Plasmodium spp. were absent in our study population. Leucocytozoon spp. have been reported in two male C. livia in South Africa in the 1990s, one from the southwestern Cape and one from Stromberg, Eastern Cape (Earle and Little 1993). The absence of Leucocytozoon spp. and Plasmodium spp. in feral pigeons has also been reported from Sao Paulo (Brazil) (Chagas et al. 2016) and Santiago (Chile) (Toro et al. 2010); however, in the latter study, no Haemoproteus spp. were found in the feral pigeon population either. Leucocytozoon spp. and Plasmodium spp. use different dipteran vectors than Haemoproteus spp. with Leucocytozoon spp. being mainly transmitted by blackflies (Simulium spp., Simuliidae, Diptera) and Plasmodium spp. by mosquitoes (Culicidae, Diptera) (Njabo et al. 2009; Russell and Mohan 1942; Ventim et al. 2012). Blackflies need running, fast-flowing streams (Rivers-Moore et al. 2007), which might be absent in highly urbanized areas in Cape Town, as suggested in a haemosporidian parasite study in black sparrowhawks (Suri et al. 2017). In that study, prevalence of Leucocytozoon spp. significantly decreased with a higher degree of urbanization; however, the prevalence for Haemoproteus spp. was similar along the urban gradient (Suri et al. 2017). The absence of vectors seems unlikely to cause the absence of Plasmodium in this study, and feral pigeons might not be native hosts for Plasmodium parasites (but see the experimentally infected C. livia with Plasmodium (Giovannolaia) gabaldoni and Plasmodium (Novyella) columbae) (Valkiūnas 2004). Generally, Plasmodium species were reported only rarely in wild feral pigeons (see exceptions, i.e., Gupta et al. 2011; Natala et al. 2009; Opara et al. 2012), but none of these records was confirmed by molecular screening methods and Haemoproteus and Plasmodium can easily be confused depending on the development stage of the parasite and the quality of the blood slide (Valkiūnas 2004).
Lastly, no effect of eumelanism on the infection intensity was found. Higher levels of eumelanism are hypothesized to have beneficial effects on the immune system (Ducrest et al. 2008) and have been shown to be correlated with improved immunocompetence in other species (Baeckens and Van Damme 2018; Figuerola et al. 1999; Galeotti and Sacchi 2003; Galván et al. 2010; Roulin et al. 2001). Lei et al. (2013) found for the color-polymorphic black sparrowhawk that darker individuals showed lower infection intensity. In the barn owl Tyto alba, individuals with more melanistic spots in their plumage had a smaller bursa of Fabricius, suggesting that they were less parasitized (Roulin et al. 2001). In feral pigeons, darker individuals are assumed to show stronger immune response, but blood parasite intensity was independent of melanism in a study by Jacquin et al. (2011), a result that is in line with our finding. Differences of blood parasite intensity and melanism were found only along an urban gradient, and only when completely white individuals were considered (Jacquin et al. 2013), which were largely absent in our study system.
To conclude, our study represents the largest dataset to date that combines sequences of mitochondrial cytochrome b (cytb) of haemosporidian parasites in feral pigeons, and provides evidence for a negative association between host body condition and parasite infection with Haemoproteus spp. However, it would be interesting to understand the long-term fitness consequences of infection by Haemoproteus spp. on feral pigeons, a species that is closely associated with humans, and has been introduced to several cities across the globe. This is especially the case, given that there is the potential for it to introduce its parasites to naïve native avifauna.
Data availability
The dataset is available as electronic supplement (Appendix Table S1), and cytochrome b sequences are available on NCBI GenBank under the accession numbers MN065190–MN065419.
References
Abramoff MD, Magalhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophoton Int 11(7):26–42
Abs M (1983) Physiology and behaviour of the pigeon. Academic Press London, London
Alizon S, de Roode JC, Michalakis Y (2013) Multiple infections and the evolution of virulence. Ecol Lett 16(4):556–567
Asghar M, Hasselquist D, Bensch S (2011) Are chronic avian haemosporidian infections costly in wild birds? J Avian Biol 42(6):530–537
Asghar M, Westerdahl H, Zehtindjiev P, Ilieva M, Hasselquist D, Bensch S (2012) Primary peak and chronic malaria infection levels are correlated in experimentally infected great reed warblers. Parasitol Int 139(10):1246–1252
Ashford RW (1971) Blood parasites and migratory fat at Lake Chad. Ibis 113(1):100–101
Baeckens S, Van Damme R (2018) Immunocompetence and parasite infestation in a melanistic and normally-coloured population of the lacertid lizard, Podarcis siculus. Amphibia-Reptilia 39(4):471–478
Beadell JS et al (2004) Prevalence and differential host-specificity of two avian blood parasite genera in the Australo-Papuan region. Mol Ecol 13(12):3829–3844
Bennett GF, Caines JR, Bishop MA (1988) Influence of blood parasites on the body-mass of passeriform birds. J Wildl Dis 24(2):339–343
Bennett GF, Peirce MA, Ashford RW (1993) Avian haematozoa: mortality and pathogenicity. J Nat Hist 27(5):993–1001
Bensch S, Hellgren O, Pérez-Tris J (2009) MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol Ecol Resour 9(5):1353–1358
Blasco R et al (2014) The earliest pigeon fanciers. Sci Rep 4:5971
Brooke RK (1981) The feral pigeon: a ‘new’ bird for the South African list. Bokmakierie 33:37–40
Chagas CRF et al (2016) Hemosporidian parasites of free-living birds in the São Paulo Zoo, Brazil. Parasitol Res 115(4):1443–1452
Chastel O, Weimerskirch H, Jouventin P (1995) Influence of body condition on reproductive decision and reproductive success in the blue petrel. Auk 112(4):964–972
Cichoiń M, Olejniczak P, Gustafsson L (1998) The effect of body condition on the cost of reproduction in female collared flycatchers Ficedula albicollis. Ibis 140(1):128–130
Clark NJ, Wells K, Dimitrov D, Clegg SM (2016) Co-infections and environmental conditions drive the distributions of blood parasites in wild birds. J Anim Ecol 85(6):1461–1470
Corbel H et al (2016) Stress response varies with plumage colour and local habitat in feral pigeons. J Ornithol 157(3):825–837
Dawson RD, Bortolotti GR (2000) Effects of hematozoan parasites on condition and return rates of American kestrels. Auk 117(2):373–380
Dean WRJ (2000) Alien birds in southern Africa: what factors determine success? S Afr J Sci 96(1):9–14
Dimitrov D, Zehtindjiev P, Bensch S, Ilieva M, Iezhova T, Valkiūnas G (2014) Two new species of Haemoproteus Kruse, 1890 (Haemosporida, Haemoproteidae) from European birds, with emphasis on DNA barcoding for detection of haemosporidians in wildlife. Syst Parasitol 87(2):135–151
Dranzoa C, Ocaido M, Katete P (1999) The ecto-, gastro-intestinal and haemo-parasites of live pigeons (Columba livia) in Kampala, Uganda. Avian Pathol 28(2):119–124
Drovetski SV et al (2014) Does the niche breadth or trade-off hypothesis explain the abundance–occupancy relationship in avian Haemosporidia? Mol Ecol 23(13):3322–3329
Ducrest A-L, Keller L, Roulin A (2008) Pleiotropy in the melanocortin system, coloration and behavioural syndromes. Trends Ecol Evol 23(9):502–510
Dunn JC et al (2017) High rates of infection by blood parasites during the nestling phase in UK Columbids with notes on ecological associations. Parasitol Int 144(5):622–628
Earle RA, Little RM (1993) Haematozoa of feral rock doves and rock pigeons in mixed flocks. S Afr J Wildl Res 23:98–98
Earle RA, Bastianello SS, Bennett GF, Krecek RC (1993) Histopathology and morphology of the tissue stages of Haemoproteus columbae causing mortality in Columbiformes. Avian Pathol 22(1):67–80
Fallon SM, Ricklefs RE (2008) Parasitemia in PCR-detected Plasmodium and Haemoproteus infections in birds. J Avian Biol 39(5):514–522
Fallon SM, Ricklefs RE, Swanson B, Bermingham E (2003) Detecting avian malaria: an improved polymerase chain reaction diagnostic. J Parasitol 89:1044–1047
Figuerola J, Munoz E, Gutiérrez R, Ferrer D (1999) Blood parasites, leucocytes and plumage brightness in the cirl bunting, Emberiza cirlus. Funct Ecol 13(5):594–601
Foronda P, Valladares B, Rivera-Medina JA, Figueruelo E, Abreu N, Casanova J (2004) Parasites of Columba livia (Aves: Columbiformes) in Tenerife (Canary Islands) and their role in the conservation biology of the laurel pigeons. Parasite 11(3):311–316
Galen SC et al (2018) The polyphyly of Plasmodium: comprehensive phylogenetic analyses of the malaria parasites (order Haemosporida) reveal widespread taxonomic conflict. R Soc Open Sci 5(5):171780
Galeotti P, Sacchi R (2003) Differential parasitaemia in the tawny owl (Strix aluco): effects of colour morph and habitat. J Zool 261(1):91–99
Galván I et al (2010) Antioxidant machinery differs between melanic and light nestlings of two polymorphic raptors. PLoS One 5(10):e13369
Garamszegi LZ (2010) The sensitivity of microscopy and PCR-based detection methods affecting estimates of prevalence of blood parasites in birds. J Parasitol 96(6):1197–1204
García-Berthou E (2001) On the misuse of residuals in ecology: testing regression residuals vs. the analysis of covariance. J Anim Ecol 70(4):708–711
Granthon C, Williams DA (2017) Avian malaria, body condition, and blood parameters in four species of songbirds. Wilson J Ornithol 129(3):492–508
Gupta DK, Jahan N, Gupta N (2011) New records of Haemoproteus and Plasmodium (Sporozoa: Haemosporida) of rock pigeon (Columba livia) in India. J Parasit Dis 35(2):155–168
Haase E, Ito S, Sell A, Wakamatsu K (1992) Melanin concentrations in feathers from wild and domestic pigeons. J Hered 83(1):64–67
Hall TA BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symposium series 1999. vol 41. [London]: Information Retrieval Ltd., c1979–c2000, pp. 95–98
Harbison CW, Jacobsen MV, Clayton DH (2009) A hitchhiker’s guide to parasite transmission: the phoretic behaviour of feather lice. Int J Parasitol 39(5):569–575
Harl J, Himmel T, Valkiūnas G, Weissenböck H (2019) The nuclear 18S ribosomal DNAs of avian haemosporidian parasites. Malar J 18(1):1–19
Hedblom M, Murgui E (2017) Urban bird research in a global perspective. In Ecology and conservation of birds in urban environments. Springer, Berlin, pp. 3–10
Huxley J (1955) Morphism in birds. Acta Int Congr Ornithol 11:309–328
Jacquin L, Lenouvel P, Haussy C, Ducatez S, Gasparini J (2011) Melanin-based coloration is related to parasite intensity and cellular immune response in an urban free living bird: the feral pigeon Columba livia. J Avian Biol 42(1):11–15
Jacquin L et al (2013) A potential role for parasites in the maintenance of color polymorphism in urban birds. Oecologia 173(3):1089–1099
Jenkins AR (2000) Hunting mode and success of African Peregrines Falco peregrinus minor, does nesting habitat quality affect foraging efficiency? Ibis 142(2):235–246
Johnston RF, Janiga M (1995) Feral pigeons, vol 4. Oxford University Press
Karamba KI, Kawo AH, Dabo NT, Mukhtar MD (2012) A survey of avian malaria parasite in Kano State, Northern Nigeria. Int J Biotechnol Mol Biol Res 3(1):8–14
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780
Kautz JE, Seamans TW (1986) Estimating age of nestling and juvenile feral rock doves. J Wildl Manag:544–547
Klopfleisch R, Werner O, Mundt E, Harder T, Teifke JP (2006) Neurotropism of highly pathogenic avian influenza virus A/chicken/Indonesia/2003 (H5N1) in experimentally infected pigeons (Columbia livia f. domestica). Vet Pathol 43(4):463–470
Knisley JO, Herman CM (1967) Haemoproteus, a blood parasite, in domestic pigeons and mourning doves in Maryland. Chesap Sci 8(3)
Korpimäki E, Tolonen P, Bennett GF (1995) Blood parasites, sexual selection and reproductive success of European kestrels. Ecoscience 2(4):335–343
Lee-Cruz L, Cunningham AA, Martínez P, Cruz M, Goodman SJ, Hamer KC (2016) Prevalence of Haemoproteus sp. in Galápagos blue-footed boobies: effects on health and reproduction. Parasitology Open 2:e1
Lei B, Amar A, Koeslag A, et al (2013) Differential haemoparasite intensity between black sparrowhawk (Accipiter melanoleucus) morphs suggests an adaptive function for polymorphism. PLoS One 8:e81607. https://doi.org/10.1371/journal.pone.0081607
Levin II et al (2011) Hippoboscid-transmitted Haemoproteus parasites (Haemosporida) infect Galapagos Pelecaniform birds: evidence from molecular and morphological studies, with a description of Haemoproteus iwa. Int J Parasitol 41(10):1019–1027
Levin II, Valkiūnas G, Iezhova TA, O’Brien SL, Parker PG (2012) Novel Haemoproteus species (Haemosporida: Haemoproteidae) from the swallow-tailed gull (Lariidae), with remarks on the host range of hippoboscid-transmitted avian hemoproteids. J Parasitol 98:847–854
Little RM (1994) Marked dietary differences between sympatric feral rock doves and rock pigeons. S Afr J Zool 29(1):33–35
Lutz HL et al (2015) Parasite prevalence corresponds to host life history in a diverse assemblage of Afrotropical birds and haemosporidian parasites. PLoS One 10(4):e0121254
Markus MB, Oosthuizen JH (1972) Pathogenicity of Haemoproteus columbae. Trans R Soc Trop Med Hyg 66(1):186–187
Martin RO, Sebele L, Koeslag A, Curtis O, Abadi F, Amar A (2014) Phenological shifts assist colonisation of a novel environment in a range-expanding raptor. Oikos 123(12):1457–1468
Marzal A, de Lope F, Navarro C, Moller AP (2005) Malarial parasites decrease reproductive success: an experimental study in a passerine bird. Oecologia 142(4):541–545
Marzal A, Bensch S, Reviriego M, Balbontin J, de Lope F (2008) Effects of malaria double infection in birds: one plus one is not two. J Evol Biol 21(4):979–987
Møller AP, Nielsen JT (2007) Malaria and risk of predation: a comparative study of birds. Ecology 88(4):871–881
Msoffe PLM, Muhairwa AP, Chiwanga GH, Kassuku AA (2010) A study of ecto-and endo-parasites of domestic pigeons in Morogoro Municipality, Tanzania. Afr J Agric Res 5(3):264–267
Mushi EZ, Binta MG, Chabo RG, Ndebele R, Panzirah R (2000) Parasites of domestic pigeons (Columba livia domestica) in Sebele, Gaborone, Botswana. J S Afr Vet Assoc 71(4):249–250
Myers N, Mittermeier RA, Mittermeier CG, Da Fonseca GA, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403(6772):853
Natala AJ et al (2009) A survey of parasites of domesticated pigeon (Columba livia domestic) in Zaria, Nigeria. Int J Soft Comput 4(4):148–150
Nebel C, Sumasgutner P, Pajot A, Amar A (2019) Response time of an avian prey to a simulated hawk attack is slower in darker conditions but is independent of hawk colour morph. R Soc Open Sci 6:190677
Njabo KY et al (2009) Coquillettidia (Culicidae, Diptera) mosquitoes are natural vectors of avian malaria in Africa. Malar J 8(1):193
Njabo KY et al (2011) Nonspecific patterns of vector, host and avian malaria parasite associations in a central African rainforest. Mol Ecol 20(5):1049–1061
Olmstead AD et al (2019) A systematic, deep sequencing-based methodology for identification of mixed-genotype hepatitis C virus infections. Infect Genet Evol 69:76–84
Opara MN, Ogbuewu IP, Iwuji CT, Ihesie EK, Etuk IF (2012) Blood characteristics, microbial and gastrointestinal parasites of street pigeons (Columbia livia) in Owerri, Imo State, Nigeria. Sci J Anim Sci 1(1):14–21
Owolabi YH, Jato ID, Yusuf KH (2009) A survey of parasites of domesticated pigeon (Columba livia domestic) in Zaria, Nigeria. Int J Soft Comput 4(4):148–150
Pacheco MA et al (2017) Mode and rate of evolution of haemosporidian mitochondrial genomes: timing the radiation of avian parasites. Mol Biol Evol 35(2):383–403
Palinauskas V, Valkiūnas G, Bolshakov CV, Bensch S (2011) Plasmodium relictum (lineage SGS1) and Plasmodium ashfordi (lineage GRW2): the effects of the co-infection on experimentally infected passerine birds. Exp Parasitol 127(2):527–533
Peirce MA, Greenwood AG, Swinnerton K (1997) Pathogenicity of Leucocytozoon marchouxi in the pink pigeon (Columba mayeri) in Mauritius. Vet Rec-Engl Edn 140(6):155–156
Pigeault R et al (2018) Haemosporidian infection and co-infection affect host survival and reproduction in wild populations of great tits. Int J Parasitol 48(14):1079–1087
Poulin R, Morand S (2000) The diversity of parasites. Q Rev Biol 75(3):277–293
Price PW et al (1986) Parasite mediation in ecological interactions. Annu Rev Ecol Syst 17(1):487–505
R Core Team (2018) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. https://www.Rproject.org
Ripley B, Venables W, Ripley MB (2016) Package ‘nnet’. R package version 7:3–12
Rivers-Moore NA, De Moor FC, Morris C, O’Keeffe J (2007) Effect of flow variability modification and hydraulics on invertebrate communities in the Great Fish River (Eastern Cape province, South Africa), with particular reference to critical hydraulic thresholds limiting larval densities of Simulium chutteri Lewis (Diptera, Simuliidae). River Res Appl 23(2):201–222
Rose S, Sumasgutner P, Koeslag A, Amar A (2017) Does seasonal decline in breeding performance differ for an African raptor across an urbanization gradient? Front Ecol Evol 5:47
Roulin A, Riols C, Dijkstra C, Ducrest A-L (2001) Female plumage spottiness signals parasite resistance in the barn owl (Tyto alba). Behav Ecol 12(1):103–110
Russell PF, Mohan BN (1942) Some mosquito hosts to avian plasmodia with special reference to Plasmodium gallinaceum. J Parasitol 28(2):127–129
Sacchi R, Gentilli A, Razzetti E, Barbieri F (2002) Effects of building features on density and flock distribution of feral pigeons Columba livia var. domestica in an urban environment. Can J Zool 80(1):48–54
Shapiro MD, Domyan ET (2013) Domestic pigeons. Curr Biol 23(8):R302–R303
Silva-Iturriza A, Ketmaier V, Tiedemann R (2012) Prevalence of avian haemosporidian parasites and their host fidelity in the central Philippine islands. Parasitol Int 61(4):650–657
Smith RJ (1999) Statistics of sexual size dimorphism. J Hum Evol 36(4):423–458
Sol D, Jovani R, Torres J (2000) Geographical variation in blood parasites in feral pigeons: the role of vectors. Ecography 23(3):307–314
Sol D, Jovani R, Torres J (2003) Parasite mediated mortality and host immune response explain age-related differences in blood parasitism in birds. Oecologia 135(4):542–547
Suri J, Sumasgutner P, Hellard É, Koeslag A, Amar A (2017) Stability in prey abundance may buffer black sparrowhawks Accipiter melanoleucus from health impacts of urbanization. Ibis 159(1):38–54
Swilling M (2006) Sustainability and infrastructure planning in South Africa: a Cape Town case study. Environ Urban 18(1):23–50
Swinnerton KJ, Peirce MA, Greenwood A, Chapman RE, Jones CG (2005) Prevalence of Leucocytozoon marchouxi in the endangered pink pigeon Columba mayeri. Ibis 147(4):725–737
Temple SA (1987) Do predators always capture substandard individuals disproportionately from prey populations? Ecology 68(3):669–674
Toro H, Saucedo C, Borie C, Gough RE, Alcaino H (2010) Health status of free-living pigeons in the city of Santiago. Avian Pathol 28(6):619–623
Valkiūnas G (2004) Avian malaria parasites and other haemosporidia. CRC
Valkiūnas G, Zickus T, Shapoval AP, Lezhova TA (2006) Effect of Haemoproteus belopolskyi (Haemosporida: Haemoproteidae) on body mass of the blackcap Sylvia atricapilla. J Parasitol 92(5):1123–1125
Valkiūnas G, Iezhova TA, Križanauskienė A, Palinauskas V, Sehgal RNM, Bensch S (2008) A comparative analysis of microscopy and PCR-based detection methods for blood parasites. J Parasitol 94(6):1395–1402
Valkiūnas G, Santiago-Alarcon D, Levin II, Iezhova TA, Parker PG (2010) A new Haemoproteus species (Haemosporida: Haemoproteidae) from the endemic Galapagos dove Zenaida galapagoensis, with remarks on the parasite distribution, vectors, and molecular diagnostics. J Parasitol 96(4):783–792
Valkiūnas G, Iezhova TA, Evans E, Carlson JS, Martinez-Gomez JE, Sehgal RN (2013) Two new Haemoproteus species (Haemosporida: Haemoproteidae) from columbiform birds. J Parasitol 99(3):513–521
van Riper C, van Riper SG, Goff ML, Laird M (1986) The epizootiology and ecological significance of malaria in Hawaiian land birds. Ecol Monogr 56(4):327–344
Vázquez B, Esperón F, Neves E, López J, Ballesteros C, Muñoz MJ (2010) Screening for several potential pathogens in feral pigeons (Columba livia) in Madrid. Acta Vet Scand 52(1):45
Ventim R, Ramos JA, Osório H, Lopes RJ, Pérez-Tris J, Mendes L (2012) Avian malaria infections in western European mosquitoes. Parasitol Res 111(2):637–645
Zylberberg M, Derryberry EP, Breuner CW, Macdougall-Shackleton EA, Cornelius JM, Hahn TP (2015) Haemoproteus infected birds have increased lifetime reproductive success. Parasitology 142(8):1033–1043
Acknowledgments
We thank S.C. McPherson, A. Jenkins and the 'Beachhouse' in Muizenberg for helping us obtain feral pigeons for this study. Furthermore, we are grateful to H. Chalmers and the team of Eagle Encounters for making their resources available to us and temporarily housing our pigeons. We thank our volunteers, S. McCarren and K. Walker, who assisted during the sample collection, and Dr. Tertius A. Groos from the Herd Care Veterinary Consultancy, Somerset West, for sharing his expertise on identifying haemosporidian parasites. Lastly, we thank the two anonymous reviewers. The quality of this paper greatly benefitted from their comments on an earlier version of this manuscript.
Funding
Open access funding provided by University of Vienna. This study was funded by the Department of Science and Technology National Research Foundation (DST-NRF) Centre of Excellence and a South Africa (NRF) / Sweden (STINT) science and technology research collaboration (Grant no. STINT 160909188048). CN received funding from the UCT’s International Student’s Scholarship and the William C. Anderson Memorial Award (Raptor Research Foundation), and PS received funding from the Claude Leon Foundation.
Author information
Authors and Affiliations
Contributions
Originally, the idea for this descriptive study came from JH, AA and PS. AP analyzed the blood slides and undertook the statistical analysis. JH carried out the molecular laboratory work and DNA sequence analyses. HW provided support in the preparation and interpretation of the data. The manuscript was prepared by CN, JH, AA, and PS and approved by all co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Ethical approval and consent to participate
This study was conducted under a CapeNature Permit (CN44-30-4175) and was approved by the University of Cape Town’s Science Faculty Animal Ethics Committee (Permit number 2018/v5/AA).
Additional information
Section Editor: Larissa Howe
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nebel, C., Harl, J., Pajot, A. et al. High prevalence and genetic diversity of Haemoproteus columbae (Haemosporida: Haemoproteidae) in feral pigeons Columba livia in Cape Town, South Africa. Parasitol Res 119, 447–463 (2020). https://doi.org/10.1007/s00436-019-06558-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-019-06558-6