Abstract
Purpose
BRCA1 and BRCA2 (BRCA1/2) are two major high-penetrance breast cancer predisposition genes, mutations in which can lead to high risks and early onset of breast cancer. This study was performed to comprehensively investigate the spectrum and prevalence of BRCA1/2 mutations in unselected Chinese breast cancer patients and evaluate the associations of BRCA1/2 mutations with related clinicopathological characteristics of the tumors.
Methods
By integrating microfluidic PCR-based target enrichment and next-generation sequencing, paired tumor and normal tissues from 313 unselected breast cancer patients were analyzed for both germline and somatic mutations of BRCA1/2 genes in Chinese Han population.
Results
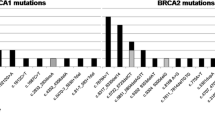
Total 5 BRCA1 and 8 BRCA2 deleterious germline mutations were detected in 5 (1.60%) and 12 (3.83%) of the 313 patients, respectively. The entire frequency of deleterious germline mutations of BRCA1/2 was 5.43%. Among them, c.1069A > T and c.3418_3419insTGACTACT in BRCA1, c.8474_8487delCATACCCTATACAG and c.6547delG in BRCA2 were novel. In addition, 32 germline variants of unknown significance in 31 (9.90%) of the 313 patients were identified. We also detected 13 somatic mutations in ten patients (3.19%), including 4 (1.28%) deleterious mutations (c.1575delT, c.2677C > T, c.7024C > T, and c.7672G > T in BRCA2) and 5 novel mutations (c.4728A > G and c.4820T > C in BRCA1; c.2527G > A, c.4069C > G and c.7672G > T in BRCA2). Notably, BRCA1 mutation carriers were significantly younger, and more likely to be ER negative and basal-like breast cancers.
Conclusions
Our study provided a reliable and effective platform for BRCA1/2 genetic testing, and suggested that there was a relatively high prevalence and special spectrum of BRCA1/2 mutations in unselected Chinese breast cancer patients.

Similar content being viewed by others

Abbreviations
- ER:
-
Estrogen receptor
- PR:
-
Progesterone receptor
- HER2:
-
Human epidermal growth factor receptor-2
- IFC:
-
Integrated fluidic circuit
- NGS:
-
Next-generation sequencing
- PARP:
-
Poly ADP-ribose polymerase
- SNV:
-
Single-nucleotide variant
- VUS:
-
Variants of unknown significance
References
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR (2010) A method and server for predicting damaging missense mutations. Nat Methods 7(4):248–249. doi:10.1038/nmeth0410-248
Al-Mulla F, Bland JM, Serratt D, Miller J, Chu C, Taylor GT (2009) Age-dependent penetrance of different germline mutations in the BRCA1 gene. J Clin Pathol 62(4):350–356. doi:10.1136/jcp.2008.062646
Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tulinius H, Thorlacius S, Eerola H, Nevanlinna H, Syrjakoski K, Kallioniemi OP, Thompson D, Evans C, Peto J, Lalloo F, Evans DG, Easton DF (2003) Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet 72(5):1117–1130. doi:10.1086/375033
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. doi:10.1093/bioinformatics/btu170
Bradbury AR, Olopade OI (2007) Genetic susceptibility to breast cancer. Rev Endocr Metab Disord 8(3):255–267. doi:10.1007/s11154-007-9038-0
Chatterjee G, Jimenez-Sainz J, Presti T, Nguyen T, Jensen RB (2016) Distinct binding of BRCA2 BRC repeats to RAD51 generates differential DNA damage sensitivity. Nucleic Acids Res. doi:10.1093/nar/gkw242
Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G (2013) Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31(3):213–219. doi:10.1038/nbt.2514
Clark SL, Rodriguez AM, Snyder RR, Hankins GD, Boehning D (2012) Structure–function of the tumor suppressor BRCA1. Comput Struct Biotechnol J. doi:10.5936/csbj.201204005
Couch FJ, Nathanson KL, Offit K (2014) Two decades after BRCA: setting paradigms in personalized cancer care and prevention. Science 343(6178):1466–1470. doi:10.1126/science.1251827
Daly MB, Pilarski R, Axilbund JE, Berry M, Buys SS, Crawford B, Farmer M, Friedman S, Garber JE, Khan S, Klein C, Kohlmann W, Kurian A, Litton JK, Madlensky L, Marcom PK, Merajver SD, Offit K, Pal T, Rana H, Reiser G, Robson ME, Shannon KM, Swisher E, Voian NC, Weitzel JN, Whelan A, Wick MJ, Wiesner GL, Dwyer M, Kumar R, Darlow S (2016) Genetic/familial high-risk assessment: breast and ovarian, version 2.2015. J Natl Compr Cancer Net 14(2):153–162
D’Andrea E, Marzuillo C, De Vito C, Di Marco M, Pitini E, Vacchio MR, Villari P (2016) Which BRCA genetic testing programs are ready for implementation in health care? A systematic review of economic evaluations. Genet Med 18(12):1171–1180. doi:10.1038/gim.2016.29
Fackenthal JD, Zhang J, Zhang B, Zheng Y, Hagos F, Burrill DR, Niu Q, Huo D, Sveen WE, Ogundiran T, Adebamowo C, Odetunde A, Falusi AG, Olopade OI (2012) High prevalence of BRCA1 and BRCA2 mutations in unselected Nigerian breast cancer patients. Int J Cancer 131(5):1114–1123. doi:10.1002/ijc.27326
Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O’Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS (2009) Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 361(2):123–134. doi:10.1056/NEJMoa0900212
Foulkes WD, Smith IE, Reis-Filho JS (2010) Triple-negative breast cancer. N Engl J Med 363(20):1938–1948. doi:10.1056/NEJMra1001389
Foulkes WD, Knoppers BM, Turnbull C (2016) Population genetic testing for cancer susceptibility: founder mutations to genomes. Nat Rev Clin Oncol 13(1):41–54. doi:10.1038/nrclinonc.2015.173
Fu Y, Jovelet C, Filleron T, Pedrero M, Motte N, Boursin Y, Luo Y, Massard C, Campone M, Levy C, Dieras V, Bachelot T, Garrabey J, Soria JC, Lacroix L, Andre F, Lefebvre C (2016) Improving the performance of somatic mutation identification by recovering circulating tumor DNA mutations. Can Res 76(20):5954–5961. doi:10.1158/0008-5472.CAN-15-3457
Gabai-Kapara E, Lahad A, Kaufman B, Friedman E, Segev S, Renbaum P, Beeri R, Gal M, Grinshpun-Cohen J, Djemal K, Mandell JB, Lee MK, Beller U, Catane R, King MC, Levy-Lahad E (2014) Population-based screening for breast and ovarian cancer risk due to BRCA1 and BRCA2. Proc Natl Acad Sci USA 111(39):14205–14210. doi:10.1073/pnas.1415979111
Gadd MA (2015) Changing beliefs and practices in women with breast cancer in China. Oncologist 20(9):979–980. doi:10.1634/theoncologist.2015-0249
Garcia C, Lyon L, Littell RD, Powell CB (2014) Comparison of risk management strategies between women testing positive for a BRCA variant of unknown significance and women with known BRCA deleterious mutations. Genet Med 16(12):896–902. doi:10.1038/gim.2014.48
Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thurlimann B, Senn HJ, Panel M (2013) Personalizing the treatment of women with early breast cancer: highlights of the St Gallen international expert consensus on the primary therapy of early breast cancer 2013. Ann Oncol 24(9):2206–2223. doi:10.1093/annonc/mdt303
Hartman AR, Kaldate RR, Sailer LM, Painter L, Grier CE, Endsley RR, Griffin M, Hamilton SA, Frye CA, Silberman MA, Wenstrup RJ, Sandbach JF (2012) Prevalence of BRCA mutations in an unselected population of triple-negative breast cancer. Cancer 118(11):2787–2795. doi:10.1002/cncr.26576
Hennessy BT, Timms KM, Carey MS, Gutin A, Meyer LA, Flake DD 2nd, Abkevich V, Potter J, Pruss D, Glenn P, Li Y, Li J, Gonzalez-Angulo AM, McCune KS, Markman M, Broaddus RR, Lanchbury JS, Lu KH, Mills GB (2010) Somatic mutations in BRCA1 and BRCA2 could expand the number of patients that benefit from poly (ADP ribose) polymerase inhibitors in ovarian cancer. J Clin Oncol 28(22):3570–3576. doi:10.1200/JCO.2009.27.2997
Hortobagyi GN, de la Garza SJ, Pritchard K, Amadori D, Haidinger R, Hudis CA, Khaled H, Liu MC, Martin M, Namer M, O’Shaughnessy JA, Shen ZZ, Albain KS, Investigators A (2005) The global breast cancer burden: variations in epidemiology and survival. Clin Breast Cancer 6(5):391–401. doi:10.3816/CBC.2005.n.043
Kais Z, Rondinelli B, Holmes A, O’Leary C, Kozono D, D’Andrea AD, Ceccaldi R (2016) FANCD2 maintains fork stability in BRCA1/2-deficient tumors and promotes alternative end-joining DNA repair. Cell Rep 15(11):2488–2499. doi:10.1016/j.celrep.2016.05.031
Koumpis C, Dimitrakakis C, Antsaklis A, Royer R, Zhang S, Narod SA, Kotsopoulos J (2011) Prevalence of BRCA1 and BRCA2 mutations in unselected breast cancer patients from Greece. Hered Cancer Clin Pract 9:10. doi:10.1186/1897-4287-9-10
Kwong A, Ng EK, Law FB, Wong HN, Wa A, Wong CL, Kurian AW, West DW, Ford JM, Ma ES (2012a) Novel BRCA1 and BRCA2 genomic rearrangements in Southern Chinese breast/ovarian cancer patients. Breast Cancer Res Treat 136(3):931–933. doi:10.1007/s10549-012-2292-1
Kwong A, Ng EK, Wong CL, Law FB, Au T, Wong HN, Kurian AW, West DW, Ford JM, Ma ES (2012b) Identification of BRCA1/2 founder mutations in Southern Chinese breast cancer patients using gene sequencing and high resolution DNA melting analysis. PLoS One 7(9):e43994. doi:10.1371/journal.pone.0043994
Lang GT, Shi JX, Hu X, Zhang CH, Shan L, Song CG, Zhuang ZG, Cao AY, Ling H, Yu KD, Li S, Sun MH, Zhou XY, Huang W, Shao ZM (2017) The spectrum of BRCA mutations and characteristics of BRCA-associated breast cancers in China: screening of 2,991 patients and 1,043 controls by next-generation sequencing. Int J Cancer 141(1):129–142. doi:10.1002/ijc.30692
Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26(5):589–595. doi:10.1093/bioinformatics/btp698
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. doi:10.1093/bioinformatics/btp352
Li Z, Guo X, Wu Y, Li S, Yan J, Peng L, Xiao Z, Wang S, Deng Z, Dai L, Yi W, Xia K, Tang L, Wang J (2015) Methylation profiling of 48 candidate genes in tumor and matched normal tissues from breast cancer patients. Breast Cancer Res Treat 149(3):767–779. doi:10.1007/s10549-015-3276-8
Maslov AY, Quispe-Tintaya W, Gorbacheva T, White RR, Vijg J (2015) High-throughput sequencing in mutation detection: a new generation of genotoxicity tests? Mutat Res 776:136–143. doi:10.1016/j.mrfmmm.2015.03.014
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20(9):1297–1303. doi:10.1101/gr.107524.110
Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W et al (1994) A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266(5182):66–71
Ng PC, Henikoff S (2001) Predicting deleterious amino acid substitutions. Genome Res 11(5):863–874. doi:10.1101/gr.176601
Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, Raine K, Jones D, Marshall J, Ramakrishna M, Shlien A, Cooke SL, Hinton J, Menzies A, Stebbings LA, Leroy C, Jia M, Rance R, Mudie LJ, Gamble SJ, Stephens PJ, McLaren S, Tarpey PS, Papaemmanuil E, Davies HR, Varela I, McBride DJ, Bignell GR, Leung K, Butler AP, Teague JW, Martin S, Jonsson G, Mariani O, Boyault S, Miron P, Fatima A, Langerod A, Aparicio SA, Tutt A, Sieuwerts AM, Borg A, Thomas G, Salomon AV, Richardson AL, Borresen-Dale AL, Futreal PA, Stratton MR, Campbell PJ, Breast Cancer Working Group of the International Cancer Genome C (2012a) The life history of 21 breast cancers. Cell 149(5):994–1007. doi:10.1016/j.cell.2012.04.023
Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, Menzies A, Martin S, Leung K, Chen L, Leroy C, Ramakrishna M, Rance R, Lau KW, Mudie LJ, Varela I, McBride DJ, Bignell GR, Cooke SL, Shlien A, Gamble J, Whitmore I, Maddison M, Tarpey PS, Davies HR, Papaemmanuil E, Stephens PJ, McLaren S, Butler AP, Teague JW, Jonsson G, Garber JE, Silver D, Miron P, Fatima A, Boyault S, Langerod A, Tutt A, Martens JW, Aparicio SA, Borg A, Salomon AV, Thomas G, Borresen-Dale AL, Richardson AL, Neuberger MS, Futreal PA, Campbell PJ, Stratton MR, Breast Cancer Working Group of the International Cancer Genome C (2012b) Mutational processes molding the genomes of 21 breast cancers. Cell 149(5):979–993. doi:10.1016/j.cell.2012.04.024
Nolan E, Vaillant F, Branstetter D, Pal B, Giner G, Whitehead L, Lok SW, Mann GB, Kathleen Cuningham Foundation Consortium for Research into Familial Breast C, Rohrbach K, Huang LY, Soriano R, Smyth GK, Dougall WC, Visvader JE, Lindeman GJ (2016) RANK ligand as a potential target for breast cancer prevention in BRCA1-mutation carriers. Nat Med 22(8):933–939. doi:10.1038/nm.4118
Paul A, Paul S (2014) The breast cancer susceptibility genes (BRCA) in breast and ovarian cancers. Front Biosci 19:605–618. doi:10.2741/4230
Plummer R (2011) Poly(ADP-ribose) polymerase inhibition: a new direction for BRCA and triple-negative breast cancer? Breast Cancer Res 13(4):218. doi:10.1186/bcr2877
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, Committee ALQA (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–424. doi:10.1038/gim.2015.30
Richter S, Haroun I, Graham TC, Eisen A, Kiss A, Warner E (2013) Variants of unknown significance in BRCA testing: impact on risk perception, worry, prevention and counseling. Ann Oncol 24:69–74. doi:10.1093/annonc/mdt312
Roy R, Chun J, Powell SN (2012) BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer 12(1):68–78. doi:10.1038/nrc3181
Sanford RA, Song J, Gutierrez-Barrera AM, Profato J, Woodson A, Litton JK, Bedrosian I, Albarracin CT, Valero V, Arun B (2015) High incidence of germline BRCA mutation in patients with ER low-positive/PR low-positive/HER-2 neu negative tumors. Cancer 121(19):3422–3427. doi:10.1002/cncr.29572
Spearman AD, Sweet K, Zhou XP, McLennan J, Couch FJ, Toland AE (2008) Clinically applicable models to characterize BRCA1 and BRCA2 variants of uncertain significance. J Clin Oncol 26(33):5393–5400. doi:10.1200/JCO.2008.17.8228
Spurdle AB, Healey S, Devereau A, Hogervorst FBL, Monteiro ANA, Nathanson KL, Radice P, Stoppa-Lyonnet D, Tavtigian S, Wappenschmidt B, Couch FJ, Goldgar DE (2012) ENIGMA-evidence-based network for the interpretation of germline mutant alleles: an international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. Hum Mutat 33(1):2–7. doi:10.1002/humu.21628
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A (2015) Global cancer statistics, 2012. CA Cancer J Clin 65(2):87–108. doi:10.3322/caac.21262
Venkitaraman AR (2002) Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108(2):171–182
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38(16):e164. doi:10.1093/nar/gkq603
Winter C, Nilsson MP, Olsson E, George AM, Chen Y, Kvist A, Torngren T, Vallon-Christersson J, Hegardt C, Hakkinen J, Jonsson G, Grabau D, Malmberg M, Kristoffersson U, Rehn M, Gruvberger-Saal SK, Larsson C, Borg A, Loman N, Saal LH (2016) Targeted sequencing of BRCA1 and BRCA2 across a large unselected breast cancer cohort suggests that one-third of mutations are somatic. Ann Oncol. doi:10.1093/annonc/mdw209
Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G (1995) Identification of the breast cancer susceptibility gene BRCA2. Nature 378(6559):789–792. doi:10.1038/378789a0
Yao L, Sun J, Zhang J, He Y, Ouyang T, Li J, Wang T, Fan Z, Fan T, Lin B, Xie Y (2016) Breast cancer risk in Chinese women with BRCA1 or BRCA2 mutations. Breast Cancer Res Treat. doi:10.1007/s10549-016-3766-3
Yates LR, Campbell PJ (2012) Evolution of the cancer genome. Nat Rev Genet 13(11):795–806. doi:10.1038/nrg3317
Zhang J, Pei R, Pang Z, Ouyang T, Li J, Wang T, Fan Z, Fan T, Lin B, Xie Y (2012) Prevalence and characterization of BRCA1 and BRCA2 germline mutations in Chinese women with familial breast cancer. Breast Cancer Res Treat 132(2):421–428. doi:10.1007/s10549-011-1596-x
Zhong X, Dong Z, Dong H, Li J, Peng Z, Deng L, Zhu X, Sun Y, Lu X, Shen F, Su X, Zhang L, Gu Y, Zheng H (2016) Prevalence and PROGNOSTIC ROLE of BRCA1/2 variants in unselected Chinese breast cancer patients. PLoS One 11(6):e0156789. doi:10.1371/journal.pone.0156789
Acknowledgements
This work was supported in part by grants from the Natural Science Foundation of China (No. 81372228), and grants from China Hunan Provincial Science and Technology Department (No. 2016GK3022, 2016XK2033, and 2016JC2068).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical standards
We declare that the experiments performed in this study comply with the current laws of the People’s Republic of China.
Conflict of interest
Xinwu Guo, Ming Chen, Xipeng Luo, Limin Peng, Xunxun Xu, and Lizhong Dai are employees of Sanway Gene Technology Inc.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, G., Guo, X., Tang, L. et al. Analysis of BRCA1/2 mutation spectrum and prevalence in unselected Chinese breast cancer patients by next-generation sequencing. J Cancer Res Clin Oncol 143, 2011–2024 (2017). https://doi.org/10.1007/s00432-017-2465-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-017-2465-8