Abstract
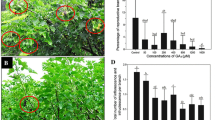
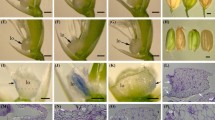
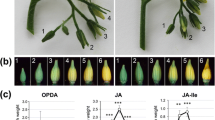
The oilseed rape plant’s transition from the vegetative to the reproductive stage is important to its yield. This transition is controlled by a large group of flowering time genes that respond to environmental and endogenous cues. The role of jasmonates in flowering is almost unknown in Brassicaceae, even in the genus Arabidopsis. In this paper, the clear effect of exogenous methyl jasmonate (MeJA) on the flowering time, floral organ morphology, and transcript levels of a group of genes implicated in floral development is shown. In controlled greenhouse experiments, we found that the effect of MeJA depended on both plant genotype and jasmonate dosage. MeJA promoted maximum flowering when it was applied to the cultivars of early flowering types of oilseed rape, such as cultivars Mei-Jian and Fu-You 4. In addition, a concentration of 100 μM resulted in the most number of early open flowers, in comparison with the results obtained for concentrations of 50 and 80 μM. Furthermore, the application of high concentrations of MeJA (100 μM) also produced various kinds of abnormal flowers. Our results demonstrated that the combined actions of the floral identity genes, specifically BnAP1, BnAP2, BnAP3, BnAG1, and BnPI3, as reflected by their respective relative transcript levels, were responsible for causing the different kinds of flower abnormalities previously undescribed in oilseed rape. We expect our assay to be an enriching addition to the body of work that attempts to understand the signaling function of jasmonates in the floral inductive pathway.








Similar content being viewed by others
Abbreviations
- AG :
-
AGAMOUSE
- AP :
-
APETALA
- COI :
-
CORONATINE INSENSITIVE
- DAD :
-
DEFECTIVE IN ANTHER DEHISCENCE
- EOIMB:
-
Early opened immature bud
- GA:
-
Gibberellins
- KNAT :
-
KNOTTED like from Arabidopsis thaliana
- MeJA:
-
Methyl jasmonate
- MYB:
-
Myeloblastosis
- OPR :
-
OPDA REDUCTASE
- PI :
-
PISTILATA
- SHA :
-
SHOOT APICAL MERISTEM ARRESTED
References
Albrechtova JTP, Ullmann L (1994) Methyl jasmonate inhibits growth and flowering in Chenopodium rubrum. Biol Plant 36:317–319
Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Barendse GWM, Croes AF, van den Ende G, Bosveld M, Creemers T (1985) Role of hormones on flower bud formation in thin-layer explant of tobacco. Biol Plant (Praha) 27:408–412
Bevan M, Walsh S (2005) The Arabidopsis genome: a foundation for plant research. Genome Res 15:1632–1642
Bowman JL, Smyth DR, Meyerowitz EM (1991) Genetic interactions among floral homeotic genes of Arabidopsis. Development 112:1–20
Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR (1993) Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119:721–743
Cheng H, Song S, Xiao L, Soo HM, Cheng Z, Xie D, Peng J (2009) Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet 5:e1000440
Ferrandiz C, Gu Q, Martienssen R, Yanofsky MF (2000) Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127:725–734
Gu Q, Ferrandiz C, Yanofsky MF, Martienssen R (1998) The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125:1509–1517
Haberer G, Mader MT, Kosarev P, Spannagl M, Yang L, Mayer KFX (2006) Large-scale cis-element detection by analysis of correlated expression and sequence conservation between Arabidopsis and Brassica oleracea. Plant Physiol 142:1589–1602
Hayama R, Coupland G (2003) Shedding light on the circadian clock and the photoperiodic control of flowering. Curr Opin Plant Biol 6:13–19
Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, Araki R, Sakurai N, Suzuki H, Aoki K, Goda H, Nishizawa OI, Shibata D, Saito K (2007) Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc Natl Acad Sci USA 104:6478–6483
Irish VF, Sussex IM (1990) Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2:741–753
Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K (2001) The DEFECTIVE IN ANTHER DEHISCIENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13:2191–2209
Ito T, Ng K, Lim T, Yu H, Meyerowitz EM (2007) The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis. Plant Cell 19:3516–3529
Jack T (2004) Molecular and genetic mechanism of floral control. Plant Cell 16:1–17
Jofuku KD, den Boer BGW, Van Montagu M, Okamuro JK (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6:1211–1225
Kempin SA, Savidge B, Yanofsky MF (1995) Molecular basis of the cauliflower phenotype in Arabidopsis. Science 267:522–525
Krajnčič B, Nemec J (1995) The effect of jasmonic acid on flowering in Spirodela polyrrhiza (L.) Schleiden. J Plant Physiol 146:754–756
Li L, Zhao Y, McCaig BC, Wingerd BA, Wang J, Whalon ME, Pichersky E, Howe GA (2004) The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16:126–143
Long Y, Shi J, Qiu D, Li R, Zhang C, Wang J, Hou J, Zhao J, Shi L, Park B-S, Choi SR, Lim YP, Meng J (2007) Flowering time quantitative trait loci analysis of oilseed Brassica in multiple environments and genomewide alignment with Arabidopsis. Genetics 177:2433–2444
Maciejewska B, Kopcewicz J (2002) Inhibitory effect of methyl jasmonate on flowering and elongation growth in Pharbitis nil. J Plant Growth Regul 21:216–223
Mandaokar A, Browser J (2009) MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiol 149:851–862
Meyerowitz EM, Bowman JL, Brockman LL, Drews GN, Jack T, Sieburth LE, Weigel D (1991) A genetic and molecular model for flower development in Arabidopsis thaliana. Development 113(Suppl 1):157–167
Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11:949–956
Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35:613–623
Osborn TC, Kole C, Parkin IAP, Shappe AG, Kuiper M, Lydiate DJ, Trick M (1997) Comparison of flowering time genes in Brassica rapa, B. napus and Arabidopsis thalianica. Genetics 146:1123–1129
Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405:200–203
Pelaz S, Gustafson-Brown C, Kohalmi SE, Crosby WL, Yanofsky MF (2001a) APETALA1 and SEPALLATA3 interact to promote flower development. Plant J 26:385–394
Pelaz S, Tapia-Lopez R, Alvarez-Buylla ER, Yanofsky MF (2001b) Conversion of leaves into petals in Arabidopsis. Curr Biol 11:182–184
Petroni K, Tonelli C, Paz-Ares J (2002) The MYB transcription factor family: from maize to Arabidopsis. Maydica 47:3–4
Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80:847–857
Quesada V, Macknight R, Dean C, Simpson GG (2003) Autoregulation of FCA pre-mRNA processing controls Arabidopsis flowering time. EMBO J 22:3142–3152
Ragni L, Belles-Boix E, Günl M, Pautot V (2008) Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis inflorescences. Plant Cell 20:888–900
Reeves PH, Coupland G (2000) Response of plant development to environment: control of flowering by daylength and temperature. Curr Opin Plant Biol 3:37–42
Sanders PM, Lee PY, Biesgen C, Boone JD, Beals TP, Weiler EW, Goldberg RB (2000) The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12:1041–1061
Schultz EA, Haughn GW (1991) LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis. Plant Cell 3:771–781
Shin B, Choi G, Yi H, Yang S, Cho I, Kim J, Lee S, Paek NC, Kim JH, Song PS, Choi G (2002) AtMYB21, a gene encoding a flower-specific transcription factor, is regulated by COP1. Plant J 30:23–32
Sonoda Y, Yao S-G, Sako K, Sato T, Kato W, Ohto M, Ichikawa K, Matsui M, Yamaguchi J, Ikeda A (2008) SHA1, a novel RING finger protein, functions in shoot apical meristem maintenance in Arabidopsis. Plant J 50:586–596
Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17:616–627
Stintzi A, Browse J (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Nat Acad Sci USA 97:10625–10630
Town CD, Cheung F, Maiti R, Crabtree J, Haas BJ, Wortman JR, Hine EE, Althoff R, Arbogast TS, Tallon LJ, Vigouroux M, Trick M, Bancroftb L (2006) Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell 18:1348–1359
Udall JA, Wendel JF (2006) Polyploidy and crop improvement. Crop Sci 46:3–13
Udvardi MK, Czechowski T, Scheible WR (2008) Eleven golden rules of quantitative RT-PCR. Plant Cell 20:1736–1737
Ueda J, Kato J (1980) Isolation and identification of a senescence promoting substance from wormwood (Artemisia absinthium L.). Plant Physiol 66:246–249
UN (1935) Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Bot 7:389–452
Wasternack C (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot 100:681–697
Wasternack C, Stenzel I, Hause B, Hause G, Kutter C, Maucher H (2006) The wound response in tomato—role of jasmonic acid. J Plant Physiol 163:297–306
Weigel D, Meyerowitz EM (1993) Activation of floral homeotic genes in Arabidopsis. Science 261:1723–1726
Weiler E (1997) Octadecanoid-mediated signal transduction in higher plants. Naturwissenschaften 84:340–349
Wilson RN, Heckman JW, Somerville C (1992) Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol 100:403–408
Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280:1091–1094
Yoshihara T, Omer E-LA, Koshino H, Sakamura S, Kikuta Y, Koda Y (1989) Structure of a tuber-inducing stimulus from potato leaves (Solanum tuberosum L.). Agric Biol Chem 53:2835–2837
Acknowledgments
The work of our laboratory was supported by Natural Science Foundation of China (abbreviated as NSFC; the program code: 30671339), National High-Tech R and D Program (abbreviated as 863 program; the program code: 2006AA10A113), and National Basic Research Program of China (abbreviated as 973 projects, program code: 2006CB101602). The authors are thankful to Prof. Ghulam Jilani, PMAS Arid Agriculture University, Pakistan, for giving critic comments; to EnPaper Company for English editing; to Institute of Agricultural and Environmental Sciences (985-IAES) for providing the convenience to use equipments.
Author information
Authors and Affiliations
Corresponding author
Additional information
H. Pak, Y. Guo contributed equally to this work.
Rights and permissions
About this article
Cite this article
Pak, H., Guo, Y., Chen, M. et al. The effect of exogenous methyl jasmonate on the flowering time, floral organ morphology, and transcript levels of a group of genes implicated in the development of oilseed rape flowers (Brassica napus L.). Planta 231, 79–91 (2009). https://doi.org/10.1007/s00425-009-1029-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-009-1029-9