Abstract
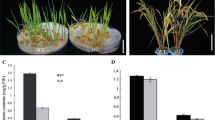
A thermo-sensitive chlorophyll deficient mutant was isolated from more than 15,000 transgenic rice lines. The mutant displayed normal phenotype at 23°C or lower temperature (permissive temperature). However, when grown at 26°C or higher (nonpermissive temperature) the plant exhibited an abnormal phenotype characterized by yellow green leaves. Genetic analysis revealed that a single nuclear-encoded recessive gene is responsible for the mutation, which is tentatively designed as cde1(t) (chlorophyll deficient 1, temporally). PCR analysis and hygromycin resistance assay indicated the mutation was not caused by T-DNA insertion. To isolate the cde1(t) gene, a map-based cloning strategy was employed and 15 new markers (five SSR and ten InDels markers) were developed. A high-resolution physical map of the chromosomal region around the cde1(t) gene was made using F2 and F3 population consisting of 1,858 mutant individuals. Finally, the cde1(t) gene was mapped in 7.5 kb region between marker ID10 and marker ID11 on chromosome 2. Sequence analysis revealed only one candidate gene, OsGluRS, in the 7.5 kb region. Cloning and sequencing of the target region from the cde1(t) mutant showed that a missense mutation occurred in the mutant. So the OsGluRS gene (TIGR locus Os02 g02860) which encode glutamyl-tRNA synthetase was identified as the Cde1(t) gene.








Similar content being viewed by others
Abbreviations
- cde1(t) :
-
Chlorophyll deficient 1, temporally
- Chl:
-
Chlorophyll
- OsGluRS :
-
Oryza glutamyl-tRNA synthetase
References
Addinall SG, Small E, Whitaker D, Sturrock S, Donachie WD, Khattar MM (2005) New temperature-sensitive alleles of ftsZ in Escherichia coli. J Bacteriol 187:358–365
Alberte RS, Hesketh JD, Hofstra G, Thornber JP, Naylor AW, Bernard RL, Brim C, Endrizzi J, Kohel RJ (1974) Composition and activity of the photosynthetic apparatus in temperature-sensitive mutants of higher plants. Proc Natl Acad Sci 71:2414–2418
Arnon DI (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol 24:1–15
Claros MG, Vincens P (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241:779–786
Eberle H, Forrest N (1982) Regulation of DNA synthesis and capacity for initiation in DNA temperature sensitive mutants of Escherichia coli. Mol Gen Genet 186:66–70
Eckhardt U, Grimm B, Hörtensteiner S (2004) Recent advances in chlorophyll biosynthesis and breakdown in higher plants. Plant Mol Biol 56:1–14
Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300:1005–1016
Goff SA Ricke D, Lan TH, Presting G, Wang RL, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, Hadley D, Hutchinson D, Martin C, Katagiri F, Lange BM, Moughamer T, Xia Y, Budworth P, Zhong JP, Miguel T, Paszkowski U, Zhang SP, Colbert M, Sun WL, Chen LL, Cooper B, Park S, Wood TC, Mao L, Quail P, Wing R, Dean R, Yu YS, Zharkikh A, Shen R, Sahasrabudhe S, Thomas A, Cannings R, Gutin A, Pruss D, Reid J, Tavtigian S, Mitchell J, Eldredge G, Scholl T, Miller RM, Bhatnagar S, Adey N, Rubano T, Tusneem N, Robinson R, Feldhaus J, Macalma T, Oliphant A, Briggs S (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296:92–100
Gálová E, Bǒhmová B, Ševčovičová A (2000) Analysis of some barley chlorophyll mutants and their response to temperature stress. Photosynthetica 38:29–35
Grossman AR, Lohr M, Im CS (2004) Chlamydomonas reinhardtii in the landscape of pigments. Annu Rev Genet 38:119–173
Hirochika H (2005) Tissue culture-induced mutations and a new type of activation tagging as tools for functional analysis of rice genes. In: Proceedings of “Rice is life: scientific perspectives for the twenty-first century”, Philippines, IRRI, pp 78–80
Hirochika H, Guiderdoni E, An G, Hsing YI, Eun MY, Han CD, Upadhyaya N, Ramachandran S, Zhang Q, Pereira A, Sundaresan V, Leung H (2004) Rice mutant resources for gene discovery. Plant Mol Biol 54:325–334
Iba K, Takamiya KI, Toh Y, Satoh H, Nishimura M (1991) Formation of functionally active chloroplasts is determined at a limited stage of leaf development in virescent mutants of rice. Dev Genet 12:342–348
International Rice Genome Sequencing Project (IRGSP) (2005) The map-based sequence of the rice genome. Nature 436:793–800
Kim YK, Lee JY, Cho HS, Lee SS, Ha HJ, Kim S, Choi D, Pai HS (2005) Inactivation of organellar glutamyl- and seryl-tRNA synthetases leads to developmental arrest of chloroplasts and mitochondria in higher plants. J Biol Chem 280:37098–37106
Kitagaki H, Ito K, Shimoi H (2004) A temperature-sensitive dcw1 mutant of Saccharomyces cerevisiae is cell cycle arrested with small buds which have aberrant cell walls. Eukaryot Cell 3:1297–1306
Kurata N, Miyoshi K, Nonomura KI, Yamazaki Y, Ito Y (2005) Rice mutants and genes related to organ development, morphogenesis and physiological traits. Plant Cell Physiol 46:48–62
Lange BM, Ghassemian M (2003) Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol Biol 51:925–948
Larkin RM, Alonso JM, Ecker JR, Chory J (2003) GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299:902–906
Lee SW, Cho BH, Park SG, Kim S (2004) Aminoacyl-tRNA synthetase complexes: beyond translation. J Cell Sci 117:3725–3734
Levican G, Katz A, Valenzuela P, Soll D, Orellana O (2005) A tRNA(Glu) that uncouples protein and tetrapyrrole biosynthesis. FEBS Lett 579:6383–6387
Luque I, Andujar A, Jia L, Zabulon G, de Marsac NT, Flores E, Houmard J (2006) Regulated expression of glutamyl-tRNA synthetase is directed by a mobile genetic element in the cyanobacterium Tolypothrix sp. PCC 7601. Mol Microbiol 60:1276–1288
Markwell J, Osterman JC (1992) Occurrence of temperature-sensitive phenotypic plasticity in chlorophyll-deficient mutants of Arabidopsis thaliana. Plant Physiol 98:392–394
Markwell JP, Danko SJ, Bauwe H, Osterman J, Gorz HJ, Haskins FA (1986) A temperature-sensitive chlorophyll b-deficient mutant of sweetclover (Melilotus alba). Plant Physiol 81:329–334
Matile P, Hörtensteiner S, Thomas H (1999) Chlorophyll degradation. Annu Rev Plant Physiol Plant Mol Biol 50:67–95
McCouch SR, Teytelman L, Xu YB, Lobos KB, Clare K, Walton M, Fu B, Maghirang R, Li Z, Xing Y, Zhang Q, Kono I, Yano M, Fjellstrom R, DeClerck G, Schneider D, Cartinhour S, Ware D, Stein L (2002) Development and mapping of 2,240 new SSR markers for rice (Oryza sativa L.). DNA Res 9:199–207
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Panaud O, Chen X, McCouch SR (1996) Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.). Mol Gen Genet 252:597–607
Park SG, Ewalt KL, Kim S (2005) Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: new perspectives on housekeepers Trends. Biochem Sci 30:569–574
Pasini L, Bruschini S, Bertoli A, Mazza R, Fracheboud Y, Marocco A (2005) Photosynthetic performance of cold-sensitive mutants of maize at low temperature. Physiol Plant 124:362–370
Pesaresi P, Varotto C, Richly E, Kurth J, Salamini F, Leister D (2001) Functional genomics of Arabidopsis photosynthesis. Plant Physiol Biochem 39:285–294
Rüdiger W (1997) Chlorophyll metabolism: from outer space down to the molecular level. Phytochemistry 46:1151–1167
Schulze JO, Masoumi A, Nickel D, Jahn M, Jahn D, Schubert WD, Heinz DW (2006) Crystal structure of a non-discriminating glutamyl-tRNA synthetase. J Mol Biol 361:888–897
Small I, Peeters N, Legeai F, Lurin C (2004) Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4:1581–1590
Strand Å (2004) Plastid-to-nucleus signalling. Curr Opin Plant Biol 7:621–625
Strand Å, Asami T, Alonso J, Ecker JR, Chory J (2003) Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421:79–83
Sugimoto H, Kusumi K, Tozawa Y, Yazaki J, Kishimoto N, Kikuchi S, Iba K (2004) The virescent-2 mutation inhibits translation of plastid transcripts for the plastid genetic system at an early stage of chloroplast differentiation. Plant Cell Physiol 45:985–996
Suzuki JY, Bollivar DW, Bauer CE (1997) Genetic analysis of chlorophyll biosynthesis. Annu Rev Genet 31:61–89
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length, variation, transposon associations, and genetic marker potential. Genome Res 11:1441–1452
Wang MB, Waterhouse PM (1997) A rapid and simple method of assaying plants transformed with hygromycin or PPT resistance genes. Plant Mol Biol Rep 15:209–215
Wang Y, Sekiguchi T, Noguchi E, Nishimoto T (2004) A hamster temperature-sensitive alanyl-tRNA synthetase mutant causes degradation of cell-cycle related proteins and apoptosis. J Biochem 135:7–16
Wellburn AR (1994) The spectral determination of chlorophyll a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J Plant Physiol 144:307–313
Yatou O, Cheng XY (1989) Temperature sensitive chlorophyll mutations. Rice Genet Newsletter 6:131
Yang X, Liang Z, Lu C (2005) Genetic engineering of the biosynthesis of glycinebetaine enhances photosynthesis against high temperature stress in transgenic tobacco plants. Plant Physiol 138:2299–2309
Yu J Hu S, Wang J, Wong GKS, Li SG, Liu B, Deng YJ, Dai L, Zhou Y, Zhang XQ, Cao ML, Liu J, Sun JD, Tang JB, Chen YJ, Huang XB, Lin W, Ye C, Tong W, Cong LJ, Geng JN, Han YJ, Li L, Li W, Hu GQ, Huang XG, Li WJ, Li J, Liu ZW, Li L, Liu JP, Qi QH, Liu JS, Li L, Li T, Wang XG, Lu H, Wu TT, Zhu M, Ni PX, Han H, Dong W, Ren XY, Feng XL, Cui P, Li XR, Wang H, Xu X, Zhai WX, Xu Z, Zhang JS, He SJ, Zhang JG, Xu JC, Zhang KL, Zheng XW, Dong JH, Zeng WY, Tao L, Ye J, Tan J, Ren XD, Chen XW, He J, Liu DF, Tian W, Tian CG, Xia HG, Bao QY, Li G, Gao H, Cao T, Wang J, Zhao WM, Li P, Chen W, Wang XD, Zhang Y, Hu JF, Wang J, Liu S, Yang J, Zhang GY, Xiong YQ, Li ZJ, Mao L, Zhou CS, Zhu Z, Chen RS, Hao BL, Zheng WM, Chen SY, Guo W, Li GJ, Liu SQ, Tao M, Wang J, Zhu LH, Yuan LP, Yang HM (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296:79–91
Zhu ZG, Xiao H, Fu YP, Hu GC, Yu YH, Si HM, Zhang JL, Sun ZX (2001) Construction of transgenic rice populations by inserting the maize transponson Ac/Ds and genetic analysis for several mutants. Chin J Biotechnol 17:288–292, (In Chinese with English abstract)
Acknowledgments
We thank Dr Qifa Zhang (Huazhong Agricultural University, China) for providing us with the pSMR-J18R plasmid. This project was supported by the grants from the National Basic Research Priorities (973) Programmes of China (G19990116-1 and 2005CB120801) and National Natural Science Foundation of China (30623006). We are grateful to Mrs Honglan Yan (China National Rice Research Institute, China) for taking pictures for the article.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, W., Fu, Y., Hu, G. et al. Identification and fine mapping of a thermo-sensitive chlorophyll deficient mutant in rice (Oryza sativa L.). Planta 226, 785–795 (2007). https://doi.org/10.1007/s00425-007-0525-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-007-0525-z