Abstract
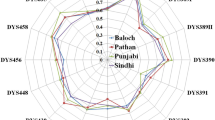
The Hazara population across Durand line has experienced extensive interaction with Central Asian and East Asian populations. Hazara individuals have typical Mongolian facial appearances and they called themselves descendants of Genghis Khan’s army. The people who speak the Balochi language are called Baloch. Previously, a worldwide analysis of Y-chromosomal haplotype diversity for rapidly mutating (RM) Y-STRs and with PowerPlex Y23 System (Promega Corporation Madison, USA) kit was created with collaborative efforts, but Baloch and Hazara population from Pakistan and Hazara population from Afghanistan were missing. In the current study, Yfiler Plus PCR Amplification Kit loci were examined in 260 unrelated Hazara individuals from Afghanistan, 153 Hazara individuals, and 111 Balochi individuals from Baluchistan Pakistan. For the Hazara population from Afghanistan and Pakistan overall, 380 different haplotypes were observed on these 27 Y-STR loci, gene diversities ranged from 0.51288 (DYS389I) to 0.9257 (DYF387S1), and haplotype diversity was 0.9992. For the Baloch population, every individual was unique at 27 Y-STR loci; gene diversity ranged from 0.5718 (DYS460) to 0.9371(DYF387S1). Twelve haplotypes were shared between 178 individuals, while only two haplotypes among these twelve were shared between 87 individuals in Hazara populations. Rst and Fst pairwise genetic distance analyses, multidimensional scaling plot, neighbor-joining tree, linear discriminatory analysis, and median-joining network were performed, which shed light on the history of Hazara and Baloch populations. The results of our study showed that the Yfiler Plus PCR Amplification Kit marker set provided substantially stronger discriminatory power in the Baloch population of Pakistan and the Hazara population across the Durand line.
Similar content being viewed by others
References
Adnan A, Ralf A, Rakha A, Kousouri N, Kayser M (2016) Improving empirical evidence on differentiating closely related men with RM Y-STRs: a comprehensive pedigree study from Pakistan. Forensic Sci Int Genet 25:45–51. https://doi.org/10.1016/j.fsigen.2016.07.005
Adnan A, Rakha A, Noor A, van Oven M, Ralf A, Kayser M (2017) Population data of 17 Y-STRs (Yfiler) from Punjabis and Kashmiris of Pakistan. Int J Legal Med. https://doi.org/10.1007/s00414-017-1611-9
Adnan A, Rakha A, Lao O, Kayser M (2018) Mutation analysis at 17 Y-STR loci (Yfiler) in father-son pairs of male pedigrees from Pakistan. Forensic Sci Int Genet. https://doi.org/10.1016/j.fsigen.2018.07.001
Vermeulen M, Wollstein A, van der Gaag K, Lao O, Xue Y, Wang Q, Roewer L, Knoblauch H, Tyler-Smith C, de Knijff P, Kayser M (2009) Improving global and regional resolution of male lineage differentiation by simple single-copy Y-chromosomal short tandem repeat polymorphisms. Forensic Sci Int Genet 3:205–213. https://doi.org/10.1016/j.fsigen.2009.01.009
Krenke BE, Viculis L, Richard ML, Prinz M, Milne SC, Ladd C, Gross AM, Gornall T, Frappier JRH, Eisenberg AJ, Barna C, Aranda XG, Adamowicz MS, Budowle B (2005) “Validation of a male-specific, 12-locus fluorescent short tandem repeat (STR) multiplex”. Forensic Sci Int 148(1): 1–14. Forensic Sci. Int. 151 (2005) 111–124. https://doi.org/10.1016/j.forsciint.2005.02.008
Mulero JJ, Chang CW, Calandro LM, Green RL, Li Y, Johnson CL, Hennessy LK (2006) Development and validation of the AmpFlSTR Yfiler PCR amplification kit: a male specific, single amplification 17 Y-STR multiplex system. J Forensic Sci 51:64–75. https://doi.org/10.1111/j.1556-4029.2005.00016.x
Thompson JM, Ewing MM, Frank WE, Pogemiller JJ, Nolde CA, Koehler DJ, Shaffer AM, Rabbach DR, Fulmer PM, Sprecher CJ, Storts DR (2013) Developmental validation of the PowerPlex® Y23 System: a single multiplex Y-STR analysis system for casework and database samples. Forensic Sci Int Genet 7:240–250. https://doi.org/10.1016/j.fsigen.2012.10.013
Kayser M, Caglià A, Corach D, Fretwell N, Gehrig C, Graziosi G, Heidorn F, Herrmann S, Herzog B, Hidding M, Honda K, Jobling M, Krawczak M, Leim K, Meuser S, Meyer E, Oesterreich W, Pandya A, Parson W, Penacino G, Perez-Lezaun A, Piccinini A, Prinz M, Schmitt C, Schneider PM, Szibor R, Teifel-Greding J, Weichhold G, de Knijff P, Roewer L (1997) Evaluation of Y-chromosomal STRs: a multicenter study. Int J Legal Med 110:125–133. https://doi.org/10.1007/s004140050051
Gopinath S, Zhong C, Nguyen V, Ge J, Lagacé RE, Short ML, Mulero JJ (2016) Developmental validation of the Yfiler ® Plus PCR Amplification Kit: an enhanced Y-STR multiplex for casework and database applications. Forensic Sci Int Genet 24:164–175. https://doi.org/10.1016/j.fsigen.2016.07.006
Jobling MA, Samara V, Pandya A, Fretwell N, Bernasconi B, Mitchell RJ, Gerelsaikhan T, Dashnyam B, Sajantila A, Salo PJ, Nakahori Y, Disteche CM, Thangaraj K, Singh L, Crawford MH, Tyler-Smith C (1996) Recurrent duplication and deletion polymorphisms on the long arm of the Y chromosome in normal males. Hum Mol Genet 5:1767–1775
Adnan A, Rakha A, Kasim K, Noor A, Nazir S, Hadi S, Pang H (2018) Genetic characterization of Y-chromosomal STRs in Hazara ethnic group of Pakistan and confirmation of DYS448 null allele. Int J Legal Med. https://doi.org/10.1007/s00414-018-1962-x
Siddique A (2012) Afghanistan’s Ethnic Divides. www.cidobafpakproject.com
Library of Congress ed. (n.d.) Afghanistan: a country study, Claitor’s Pub. Division, c2001, Baton Rouge
Dashti N (2012) The Baloch and Balochistan: a historical account from the beginning to the fall of the Baloch State, Trafford Sl
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462
Michalakis Y, Excoffier L (1996) A generic estimation of population subdivision using distances between alleles with special reference for microsatellite loci. Genetics 142:1061–1064
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. MolBiolEvol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
He G, Adnan A, Rakha A, Yeh H-Y, Wang M, Zou X, Guo J, Rehman M, Fawad A, Chen P, Wang C-C (2019) A comprehensive exploration of the genetic legacy and forensic features of Afghanistan and Pakistan Mongolian-descent Hazara. Forensic Sci Int Genet 42:e1–e12. https://doi.org/10.1016/j.fsigen.2019.06.018
Haber M, Platt DE, AshrafianBonab M, Youhanna SC, Soria-Hernanz DF, Martínez-Cruz B, Douaihy B, Ghassibe-Sabbagh M, Rafatpanah H, Ghanbari M, Whale J, Balanovsky O, Wells RS, Comas D, Tyler-Smith C, Zalloua PA (2012) The genographic consortium, Afghanistan’s ethnic groups share a Y-chromosomal heritage structured by historical events. PLoS ONE 7:e34288. https://doi.org/10.1371/journal.pone.0034288
Adnan A, He G, Rakha A, Kasimu K, Guo J, Hassan SE, Hadi S, Wang C-C, Xuan J (2019) Phylogenetic relationship and genetic history of Central Asian Kazakhs inferred from Y-chromosome and autosomal variations. Mol Genet Genomics. https://doi.org/10.1007/s00438-019-01617-0
Zerjal T, Xue Y, Bertorelle G, Wells RS, Bao W, Zhu S, Qamar R, Ayub Q, Mohyuddin A, Fu S, Li P, Yuldasheva N, Ruzibakiev R, Xu J, Shu Q, Du R, Yang H, Hurles ME, Robinson E, Gerelsaikhan T, Dashnyam B, Mehdi SQ, Tyler-Smith C (2003) The genetic legacy of the Mongols. Am J Hum Genet 72:717–721. https://doi.org/10.1086/367774
Smolenyak MA (2004) Turner, Trace your roots with DNA: using genetic tests to explore your family tree, Rodale ; Distributed to the trade by Holtzbrinck Publishers, Emmaus, Pa.: New York
Wells S (2007) Deep ancestry: inside the genographic project. Washington, D.C.: National Geographic
Basu A, Sarkar-Roy N, Majumder PP (2016) Genomic reconstruction of the history of extant populations of India reveals five distinct ancestral components and a complex structure. Proc Natl Acad Sci 113:1594–1599. https://doi.org/10.1073/pnas.1513197113
Di Cristofaro J, Pennarun E, Mazières S, Myres NM, Lin AA, Temori SA, Metspalu M, Metspalu E, Witzel M, King RJ, Underhill PA, Villems R, Chiaroni J (2013) Afghan Hindu Kush: where Eurasian sub-continent gene flows converge. PLoS ONE 8:e76748. https://doi.org/10.1371/journal.pone.0076748
Mcelreavey K, Quintana-Murci L (2005) A population genetics perspective of the Indus Valley through uniparentally-inherited markers. Ann Hum Biol 32:154–162. https://doi.org/10.1080/03014460500076223
Sengupta S, Zhivotovsky LA, King R, Mehdi SQ, Edmonds CA, Chow C-ET, Lin AA, Mitra M, Sil SK, Ramesh A, Usha Rani MV, Thakur CM, Cavalli-Sforza LL, Majumder PP, Underhill PA (2006) Polarity and temporality of high-resolution Y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet 78:202–221. https://doi.org/10.1086/499411
Qamar R, Ayub Q, Mohyuddin A, Helgason A, Mazhar K, Mansoor A, Zerjal T, Tyler-Smith C, Mehdi SQ (2002) Y-chromosomal DNA variation in Pakistan. Am J Hum Genet 70:1107–1124. https://doi.org/10.1086/339929
Tarlykov PV, Zholdybayeva EV, Akilzhanova AR, Nurkina ZM, Sabitov ZM, Rakhypbekov TK, Ramanculov EM (2013) Mitochondrial and Y-chromosomal profile of the Kazakh population from East Kazakhstan. Croat Med J 54:17–24
Chang C-W, Mulero JJ, Budowle B, Calandro LM, Hennessy LK (2006) Identification of a novel polymorphism in the X-chromosome region homologous to the DYS456 locus. J Forensic Sci 51:344–348. https://doi.org/10.1111/j.1556-4029.2006.00052.x
Redd AJ, Agellon AB, Kearney VA, Contreras VA, Karafet T, Park H, de Knijff P, Butler JM, Hammer MF (2002) Forensic value of 14 novel STRs on the human Y chromosome. Forensic Sci Int 130:97–111
Balaresque P, Bowden GR, Parkin EJ, Omran GA, Heyer E, Quintana-Murci L, Roewer L, Stoneking M, Nasidze I, Carvalho-Silva DR, Tyler-Smith C, de Knijff P, Jobling MA (2008) Dynamic nature of the proximal AZFc region of the human Y chromosome: multiple independent deletion and duplication events revealed by microsatellite analysis. Hum Mutat 29:1171–1180. https://doi.org/10.1002/humu.20757
Park MJ, Shin K-J, Kim NY, Yang WI, Cho S-H, Lee HY (2008) Characterization of deletions in the DYS385 flanking region and null alleles associated with AZFc microdeletions in Koreans. J Forensic Sci 53:331–334. https://doi.org/10.1111/j.1556-4029.2008.00660.x
Acknowledgements
We thank all volunteers who provided material and data for this project, especially Muhammad Rehman and Abulhasan Fawad.
Funding
This study was financially supported by the China Medical University postdoctoral research grant (100/1210619014).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
All participants who were included in this study were unrelated individuals of at least three generations.
Informed consent
All participants gave their informed consent either orally and with thumbprint (in case they could not write) or in writing after the study aims and procedures were carefully explained to them.
Ethical approval
This collaborative study was approved by the ethical review boards of China Medical University, Shenyang, Liaoning Province, People’s Republic of China (2019/067-P), University of Health Sciences Lahore Pakistan (2017-CMU-1/14), and Ministry of Public Health, Forensic Medicine Directorate, Kabul, Afghanistan (FC-2017–02). All the experimental procedures were performed in accordance with the standards of the Declaration of Helsinki.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
414_2021_2591_MOESM3_ESM.jpg
Supplementary Figure 2 Two-dimensional plot from multi-dimensional scaling analysis of Rst-values based on Yfiler haplotypes Ne (JPG 282 KB)
414_2021_2591_MOESM4_ESM.jpg
Supplementary Figure 3 Neighbor-joining tree based on the Fst values between the Baloch population of Pakistan and Hazara populations across the Durand line with reference populations (JPG 2228 KB)
414_2021_2591_MOESM5_ESM.jpg
Supplementary Figure 4 The median-joining network of the Baloch population of Pakistan and Hazara populations across the Durand line based on 20 Y STRs (JPG 173 KB)
414_2021_2591_MOESM7_ESM.jpg
Supplementary Figure 6 LDA Analysis between the Baloch population of Pakistan and Hazara populations across the Durand line, Central Asia, South Asia, Russia, and East Asian populations (JPG 97 KB)
414_2021_2591_MOESM11_ESM.xlsx
Supplementary Table 3 Reference Populations from Central, Eastern and South Asia populations selected as reference populations used in LDA, NJ tree and multidimensional scaling (MDS) analysis (XLSX 11 KB)
414_2021_2591_MOESM14_ESM.xlsx
Supplementary Table 6 Sequence in the relevant flanking and repeat region of the DYS448 locus for null alleles (XLSX 12 KB)
Rights and permissions
About this article
Cite this article
Adnan, A., Rakha, A., Nazir, S. et al. Forensic features and genetic legacy of the Baloch population of Pakistan and the Hazara population across Durand line revealed by Y-chromosomal STRs. Int J Legal Med 135, 1777–1784 (2021). https://doi.org/10.1007/s00414-021-02591-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-021-02591-2