Abstract
Key message
PcWRKY33 is a transcription factor which can reduce salt tolerance by decreasing the expression of stress-related genes and increasing the cellular levels of reactive oxygen species (ROS).
Abstract
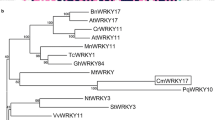
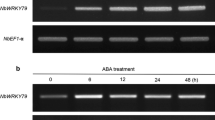
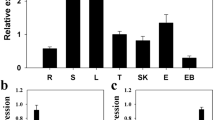
WRKY transcription factors play important roles in the regulation of biotic and abiotic stresses. Here, we report a group I WRKY gene from Polygonum cuspidatum, PcWRKY33, that encodes a nucleoprotein, which specifically binds to the W-box in the promoter of target genes to regulate their expression. The results from qPCR and promoter analysis show that expression of PcWRKY33 can be induced by various abiotic stresses, including NaCl and plant hormones. Overexpression of PcWRKY33 in Arabidopsis thaliana reduced tolerance to salt stress. More specifically, several physiological parameters (such as root length, seed germination rate, seedling survival rate, and chlorophyll concentration) of the transgenic lines were significantly lower than those of the wild type under salt stress. In addition, following exposure to salt stress, transgenic plants showed decreased expression of stress-related genes, a weakened ability to maintain Na+/K+ homeostasis, decreased activities of reactive oxygen species- (ROS-) scavenging enzymes, and increased accumulation of ROS. Taken together, these results suggest that PcWRKY33 negatively regulates the salt tolerance in at least two ways: by down-regulating the induction of stress-related genes and by increasing the level of cellular ROS. In sum, our results indicate that PcWRKY33 is a group I WRKY transcription factor involved in abiotic stress regulation.






Similar content being viewed by others
References
Agarwal SK, Singh SS, Verma S, Kumar S (2000) Antifungal activity of anthraquinone derivatives from Rheum emodi. J Ethnopharmacol 72:43–46
Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285:1256
Barnard DL, Fairbairn DW, O’Neill KL, Gage TL, Sidwell RW (1995) Anti-human cytomegalovirus activity and toxicity of sulfonated anthraquinones and anthraquinone derivatives. Antivir Res 28:317–329
Bartels D, Sunkar R (2005) Drought and salt tolerance in plants. Crit Rev Plant Sci 24:23–58
Bhattacharjee S (2005) Reactive oxygen species and oxidative burst: roles in stress, senescence and signal transduction in plants. Curr Sci India 89:1113–1121
Chao D, Zhao P, Zhang H, Li N, Zheng L, Wang Y (2017) The Reaumuria trigyna transcription factor RtWRKY1 confers tolerance to salt stress in transgenic Arabidopsis. J Plant Physiol 215:48–58
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cui MH et al (2013) An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett 587:1773–1778
Devaiah BN, Karthikeyan AS, Raghothama KG (2007) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143:1789–1801
Duan Y et al (2015) PtrWRKY73, a salicylic acid-inducible poplar WRKY transcription factor, is involved in disease resistance in Arabidopsis thaliana. Plant Cell Rep 34:831–841
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5:199–206. https://doi.org/10.1016/S1360-1385(00)01600-9
FAO (2005) Global network on integrated soil management for sustainable use of salt-affected soils. Rome, Italy: FAO Land and Plant Nutrition Management Service. http://www.fao.org/ag/agl/agll/spush
Fu QT, Yu DQ (2010) Expression profiles of AtWRKY25, AtWRKY26 and AtWRKY33 under abiotic stresses. Hereditas 32:848–856
Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Bioch 48:909–930
Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Phys 51:463–499
Hu W et al (2013) TaASR1, a transcription factor gene in wheat, confers drought stress tolerance in transgenic tobacco. Plant Cell Environ 36:1449–1464
Jiang Y, Deyholos MK (2009) Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol Biol 69:91–105
Kato N et al (2007) Identification of a WRKY protein as a transcriptional regulator of benzylisoquinoline alkaloid biosynthesis in Coptis japonica. Plant Cell Physiol 48:8–18
Li J, Brader G, Palva ET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16:319–331
Li HX et al (2013) Cerebroside C increases tolerance to chilling injury and alters lipid composition in wheat roots. Plos One 8:e73380
Li P et al (2015) Chrysanthemum WRKY gene CmWRKY17 negatively regulates salt stress tolerance in transgenic chrysanthemum and Arabidopsis plants. Plant Cell Rep 34:1365–1378
Liang QY et al (2017) Chrysanthemum WRKY gene DgWRKY5 enhances tolerance to salt stress in transgenic chrysanthemum. Sci Rep 7:4799
Lichtenthaler HK, Wellburn AR (1983) Determination of total carotenoids and chlorophylls a and b of leaf in different solvents. Biochem Soc Trans 11:591–592
Liu QL, Zhong M, Li S, Pan YZ, Jiang BB, Jia Y, Zhang HQ (2013) Overexpression of a chrysanthemum transcription factor gene, DgWRKY3, in tobacco enhances tolerance to salt stress. Plant Physiol Bioch 69:27–33
Liu QL, Xu KD, Pan YZ, Jiang BB, Liu GL, Jia Y, Zhang HQ (2014) Functional analysis of a novel chrysanthemum WRKY transcription factor gene involved in salt tolerance. Plant Mol Biol Rep 32:282–289
Liu X, Song Y, Xing F, Wang N, Wen F, Zhu C (2016) GhWRKY25, a group I WRKY gene from cotton, confers differential tolerance to abiotic and biotic stresses in transgenic Nicotiana benthamiana. Protoplasma 253:1265–1281
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Mahajan S, Tuteja N (2005) Cold, salinity and drought stresses: an overview. Arch Biochem Biophys 444:139–158
Matsui A et al (2008) Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol 49:1135–1149
Merz PR, Moser T, Höll J, Kortekamp A, Buchholz G, Zyprian E, Bogs J (2015) The transcription factor VvWRKY33 is involved in the regulation of grapevine (Vitis vinifera) defense against the oomycete pathogen Plasmopara viticola. Physiol Plant 153:365–380
Miao Y, Laun T, Zimmermann P, Zentgraf U (2004) Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol 55:853–867
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7:405–410
Moore K, Roberts LJ (1998) Measurement of lipid peroxidation. Free Radic Res 28:659–671
Rushton PJ, Somssich IE (1998) Transcriptional control of plant genes responsive to pathogens. Curr Opin Plant Biol 1:311–315
Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE (1996) Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J 15:5690–5700
Rushton PJ et al (2008) Tobacco transcription factors: novel insights into transcriptional regulation in the solanaceae. Plant Physiol 147:280–295
Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. PNAS 97:6896–6901
Shi J, An HL, Zhang L, Gao Z, Guo XQ (2010) GhMPK7, a novel multiple stress-responsive cotton group C MAPK gene, has a role in broad spectrum disease resistance and plant development. Plant Mol Biol 74:1–17
Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA (1995) An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res 23:1087–1088
Su YT, Chang HL, Shyue SK, Hsu SL (2005) Emodin induces apoptosis in human lung adenocarcinoma cells through a reactive oxygen species-dependent mitochondrial signaling pathway. Biochem Pharmacol 70:229–241
Ulker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7:491–498
Ullah A, Sun H, Yang X, Zhang X (2017) A novel cotton WRKY-gene, GhWRKY6-like, improves salt tolerance by activating the ABA signalling pathway and scavenging of reactive oxygen species. Physiol Plant. https://doi.org/10.1111/ppl.12651
Wang Y, Shu Z, Wang W, Jiang X, Li D, Pan J, Li X (2016) CsWRKY2, a novel WRKY gene from Camellia sinensis, is involved in cold and drought stress responses. Biol Plantarum 60:1–9
Wang K et al (2017) Overexpression of DgWRKY4 enhances salt tolerance in chrysanthemum seedlings. Front Plant Sci 8:1592. https://doi.org/10.3389/fpls.2017.01592
Xu Z, Wang C, Xue F, Zhang H, Ji W (2015) Wheat NAC transcription factor TaNAC29 is involved in response to salt stress. Plant Physiol Bioch 96:356–363
Yan H, Jia H, Chen X, Hao L, An H, Guo X (2014) The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol 55:2060–2076
Yang YO, Li RG, Min Q (2010) In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J 22:543–551
Yen GC, Duh PD, Chuang DY (2000) Antioxidant activity of anthraquinones and anthrone. Food Chem 70:437–441
Zhang Y, Wang L (2005) The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol Biol 5:1
Zhou QY et al (2008) Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J 6:486–503. https://doi.org/10.1111/j.1467-7652.2008.00336.x
Zhou J, Li F, Wang JL, Ma Y, Chong K, Xu YY (2009) Basic helix-loop-helix transcription factor from wild rice (OrbHLH2) improves tolerance to salt- and osmotic stress in Arabidopsis. J Plant Physiol 166:1296–1306
Zhou L, Wang NN, Gong SY, Lu R, Li Y, Li XB (2015) Overexpression of a cotton (Gossypium hirsutum) WRKY gene, GhWRKY34, in Arabidopsis enhances salt-tolerance of the transgenic plants. Plant Physiol Bioch 96:311–320
Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6:66–71
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Zhu JK (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6:441–445
Zou C, Jiang W, Yu D (2010) Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. J Exp Bot 61:3901–3914
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No.61672489; 61379081).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest regarding the publication of this paper.
Additional information
Communicated by Kang Chong.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bao, W., Wang, X., Chen, M. et al. A WRKY transcription factor, PcWRKY33, from Polygonum cuspidatum reduces salt tolerance in transgenic Arabidopsis thaliana. Plant Cell Rep 37, 1033–1048 (2018). https://doi.org/10.1007/s00299-018-2289-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-018-2289-2