Abstract
Hexaploid wheat, one of the world’s most important staple crops, remains a challenge for genetic transformation. We are developing a floral transformation protocol for wheat that does not require tissue culture. This paper presents three transformants in the hard red germplasm line Crocus that have been characterized thoroughly at the molecular level over three to six generations. Wheat spikes at the early boot stage, i.e. the early, mid or late uninucleate microspore stages, were immersed in an infiltration medium of strain C58C1 harboring pDs(Hyg)35S, or strain AGL1 harboring pBECKSred. pDs(Hyg)35S contains the NPTII and hph selectable markers, and transformants were detected using paromomycin spray at the whole plant level, NPTII ELISAs, or selection on medium with hygromycin. Strain AGL1, harboring pBECKSred, which contains the maize anthocyanin regulators, Lc and C1, and the NPTII gene, was also used to produce a Crocus transformant. T1 and T2 seeds with red embryos were selected; T1 and T2 plants were screened by sequential tests for paromomycin resistance and NPTII ELISAs. The transformants were low copy number and showed Mendelian segregation in the T2. Stable transmission of the transgenes over several generations has been demonstrated using Southern analysis. Gene expression in advanced progeny was shown using Reverse Transcriptase-PCR and ELISA assays for NPTII protein expression. This protocol has the potential to reduce the time and expense required for wheat transformation.
Similar content being viewed by others
Introduction
Genetic transformation is an essential tool for analyzing gene function in plants. Most published protocols for the transformation of hexaploid wheat (Triticum aestivum L.) involve the use of tissue culture, skilled personnel and specialized equipment that may not be available to all researchers, especially those in developing countries. Currently, transgenes are typically introduced using particle bombardment (biolistics) and Agrobacterium-mediated transformation of dissected explants. Particle bombardment has been used to transform embryogenic wheat calli (Vasil et al. 1992; Weeks et al. 1993) and dissected scutella (Nehra et al. 1994), and T-DNA transfer by Agrobacterium has been achieved with embryos, pre-cultured immature embryos, and embryogenic calli (Cheng et al. 1997). These approaches rely on the totipotency of individual plant cells to dedifferentiate into unorganized callus tissue, become embryogenic and regenerate into whole plants through organogenesis. Immature embryos are primarily used because of their greater capacity to regenerate plants (Zale et al. 2004).
Cereal transformation via a tissue culture phase has been successful, but involves several limitations. The use of tissue culture allows selection of single transformed cells which are regenerated in a whole plant and this lessens the production of genetic chimeras. However, the tissue culture approach causes somaclonal variation due to either epigenetic effects or chromosomal rearrangements (Kaeppler et al. 2000; Mohan Jain 2001). For example, many of the two hundred thousand T-DNA lines produced by tissue culture in rice are somaclonal variants (An et al. 2005). In the past, transformation success in wheat has been limited to a relatively few genotypes that regenerate well from tissue culture (Jones 2005; Pellegrineschi et al. 2002). Biolistics can also cause multiple T-DNA insertions and gene silencing in subsequent generations (Taylor and Fauquet 2002).
Attempts at floral transformation of allohexaploid wheat (Triticum aestivum L.) were published prior to the development of the floral dip method in Arabidopsis. The target tissue in these experiments was the pollen, and a basal medium containing Agrobacterium harboring genetic constructs was pipetted into open wheat florets at anthesis. In transformants isolated by Hess et al. (1990), the T-DNA underwent size alterations or appeared to be lost in subsequent generations. Langridge et al. (1992) concluded that floral transformation of wheat, barley and maize at anthesis led to artifacts on gels in the T0 generation possibly due to transformation of an endophytic bacterium.
In planta transformation of the model dicotyledonous species, Arabidopsis thaliana by vacuum infiltration of whole plants (Bechtold et al. 1993) and the floral dip (Clough and Bent 1998) are now routine, and have contributed greatly to the rapid forward and reverse genetics research in this species. Three different laboratories have confirmed that the target of T-DNA transfer in Arabidopsis is primarily the female ovule, and segregation data showed that the T-DNA insertions are hemizygous (Bechtold et al. 2000; Desfeux et al. 2000; Ye et al. 1999). Similar approaches have been developed for the in planta transformation a number of other dicotyledonous species such as Shepard’s purse (Capsella bursa-pastoris), radish, and alfalfa (Medicago sativa) (Bartholmes, et al. 2008; Curtis and Nam 2001; Weeks, et al. 2008).
A number of advances in reducing the dependence on tissue culture have recently been made in wheat transformation. An in planta transformation method in which an Agrobacterium coated needle is used to inoculate a germinating wheat seedling has been developed (Supartana et al. 2006). Another method involves inoculating Agrobacterium on the basal portion of cut seedlings with no intervening callus phase and requires minimal tissue culture (Zhao et al. 2006).
The objective of this research was to determine whether stable transformants of wheat could be obtained by the floral dip if the treatment were performed at an earlier stage of floral development than previously used (Hess et al. 1990; Langridge et al. 1992). This was based on the rationale that the target of transformation may be the ovule, as in Arabidopsis (Bechtold et al. 2000; Desfeux et al. 2000; Ye et al. 1999). An infiltration medium containing a surfactant and acetosyringone was used to produce multiple stable transformants in the ‘Crocus’ genotype, a hard red wheat germplasm line that possesses the double recessive alleles for high crossability with rye (Zale and Scoles 1999). In one version of the protocol, putative transformants were screened with a paromomycin spray at the whole plant level (Cheng et al. 1997) or germinated on hygromycin medium followed by NPTII ELISAs. In another version of the protocol, the embryos of T1 seeds were visually screened for color changes induced by the maize anthocyanin regulatory genes. Southern hybridizations for transgene copy number have shown that the T-DNA has integrated in the wheat genome with stable transmission of the transgenes over three to six generations. Finally, RT-PCR and NPTII ELISA assays demonstrated expression of the transgenes in advanced progenies.
Materials and methods
Plant material and growth conditions
Crocus spring wheat germplasm was obtained from the USDA/ARS National Germplasm Resources Information Network (GRIN http://www.ars-grin.gov/ ). Crocus is a hard red spring wheat germplasm line adapted to the Northern Great Plains and it also possesses the crossability alleles (Zale and Scoles 1999). The original rationale was that if the crossability alleles of ‘Crocus’ provide a decreased barrier in wide crosses, and then they might also result in a reduced barrier to Agrobacterium in the pistil. For example, these alleles are associated with decreased expression of nucleases in the carpal at anthesis (Zale, unpublished) and might increase the probability of floral transformation. Moreover, a red seeded wheat line was chosen because expression of maize Lc/C1 in white wheat lines is known to be detrimental to plant growth (McCormac et al. 1997). Zuzanna spring wheat is a Czech variety and was a gift from Dr. Ludmila Ohnoutkova, The University of Tennessee, Department of Plant Sciences.
The wheat plants were grown in the greenhouse or in walk-in growth rooms, with day temperatures at 23 ± 4°C with a 16 h light cycle; the night temperatures were 16 ± 4°C with an 8-h dark cycle. The intensity of the lighting never fell below 400 μE/m2 per second.
Vectors
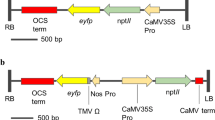
Plants were transformed with vector pDs(Hyg)35S and pBECKSred. The pDs(Hyg)35S is a binary vectors and part of a two component Ac/Ds system used for activation tagging in Arabidopsis (Long, et al. 1997; Wilson, et al. 1996). pDs(Hyg)35S contains the NPTII gene driven by the nos promoter; a maize Ds element and a hygromycin gene interrupting a spectinomycin excision marker (Fig. 1a) pBECKSred is a binary vector carrying the NPTII gene driven by nos promoter and the maize transcription factors Lc and C1 (McCormac et al. 1997). Lc is driven by the 35S promoter; C1 is driven by the 35S promoter:Adh1 intron1 (Fig. 1b).
Schematic diagrams of the T-DNA constructs used in the floral dip including rationale for Southern analysis. a Diagram of the T-DNA of pDs(Hyg)35S showing the location of the EcoRI (E1) and BglII restriction sites relative to the NPTII probe (probe 1) and Hph probe (probe 2). The position of the Ds transposable element, the neomycin phosphotransferase (NPTII) gene, the spectinomycin (SPT) resistance gene, the hygromycin (Hph) resistance gene, Left Border (LB) and Right Border (RB) are shown. b Diagram of the T-DNA of pBECKSred showing the location of the EcoRI (E1), HindIII (HIII), BamHI (B1), and Eag1 restriction sites relative to the NPTII probe (probe1) and the C1 probe (probe 2)
Agrobacterium strains
The Agrobacterium tumefaciens strains C58C1 and AGL1 were used in transformation experiments. The C58C1 strain was chosen because its relative, C58, has been used to transform monocots including wheat (Cheng et al. 1997) and dicots (Long et al. 1997; Wilson et al. 1996). AGL1 is a derivative of C58 containing a deletion in the recA gene that lessens genetic rearrangements of the Ti plasmid in Agrobacterium (Lazo et al. 1991), and is useful in monocot transformation (Pacurar et al. 2007; Wu et al. 2007).
Agrobacterium culture and infiltration medium
Five ml of LB broth was inoculated with the appropriate antibiotics and grown overnight at 22 ± 4°C with shaking at 2×g. The next day, 2.5 ml of this culture was used to inoculate 250 ml of YEP broth containing the antibiotics and 200 μM acetosyringone, and grown at 22 ± 4°C with shaking at 2×g until the OD600 = 0.8–1.0. The YEP/Agrobacterium culture was centrifuged at 6,400×g for 15 min at room temperature and the supernatant discarded. The cells were gently re-suspended in infiltration medium consisting of half-strength Murashige and Skoog (MS) (Murashige and Skoog 1962) medium (Fisher Biotech, Pittsburgh, PA), 5% w/v sucrose, 0.5 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer, pH 5.8 (Bechtold et al. 1993; Clough and Bent 1998), and 200 μM acetosyringone. The wetting agent Silwet L-77, at 0.4% (v/v) was added just prior to dipping.
Preparation of the wheat spikes and floral transformation
Preanthesis wheat spikes at the early, mid or late uninucleate microspore stage were used for dipping. This is the early boot stage (Fig. 2). Plants were well watered one day prior to dipping. The lengths of the spikes will vary in different environmental conditions, but the best results are obtained when the spikes are approximately 6–7 cm in length and have not emerged from the sheath. The spikes are gently unsheathed with a small pair of scissors and the developing spikes carefully removed. The terminal florets and the inner florets can be removed as they are frequently sterile. The remaining florets are clipped to the end of the glumes. Spikes are immersed in the Agrobacterium infiltration media for 1.5–2 min and the spikes were covered with plastic bags for two days to ensure high humidity. Afterwards, the florets are uncovered to air dry. As the spikes approach anthesis, they can be covered in glassine bags to prevent cross pollination, although this lowers seed set. Mature seeds, designated as T1, are harvested and screened for transformation events. In one instance, spikes were dipped twice (Bent 2000), i.e. they were dipped once, and then the dipping procedure was repeated the next morning.
Screening of transformants
Screening of putative transformants typically commenced in the T1 generation but never with PCR because of its sensitivity of to bacterial contamination and the preponderance of positive plants. Seeds derived from T0 plants treated with pBECKSred, were visually examined under a stereoscope and those with red embryos (see Fig. S1) were selected for further testing. Seeds were washed in 30–70% commercial bleach for 30 min with shaking, rinsed three times in sterile distilled H2O, 5 min per wash, and planted in 6 in. pots containing potting mix.
A series of sequential screens were then used to identify putatively transformed T1 plants containing pDs(Hyg)35S or pBECKSred. Screening for expression of the NPTII gene at the whole plant level was performed by spraying a 2% paromomycin solution containing 0.02% Tween 20 on seedlings at the two–three leaf stage and scoring for minimal bleaching 7–10 days later (Cheng et al. 1997). Tissue was harvested from paromomycin-screened plants and tests for NPTII protein expression by ELISA assay performed according to manufacturer’s instructions (Agdia, Elkhart, Indiana). Positive plants were saved if ELISA readings were greater than those of wild-type Crocus germplasm.
Segregation ratios and statistical analyses
Segregation of T2 progeny derived from independent transformation events was determined using 2% paromomycin at the whole plant level (Cheng et al. 1997), NPTII ELISAs, growth on hygromycin, or scoring for seed color changes induced by the maize anthocyanin regulatory genes. The segregation ratios were subjected to χ2 Goodness of Fit tests (Mead et al. 2003).
Genomic DNA digestions, probes and Southern blots
Genomic DNA was extracted with either using the DNAzol™ procedure (Invitrogen, Carlsbad, CA) or the CTAB DNA extraction procedure (Kleinhofs et al. 1993). Genomic DNA (25 µg) was digested to completion with restriction enzymes (New England Biolabs, Ipswich, MA), and blotted onto Gene Screen™ membrane (Perkin Elmer NEN, Waltham, MA) or Hybond N+™ (Amersham Pharmacia, Piscataway, NJ) using the alkaline transfer procedure (Sambrook and Russell 2001). The rapid downward Southern blot (Chomczynski 1993) method was employed to improve transfer of the DNA to the membrane. Left border analysis was performed to determine copy number (Cheng et al. 1997), and transgene analysis performed using enzymes that cut on either side of the transgene (Jordan 2000).
The NPTII or hph probes used in Southern hybridizations were PCR amplified. Gene specific primers to the NPTII sequence were used to amplify a 929 bp product from pBECKSred using the NPTII Forward 5′-GCTTGGGTGGAGAGGCTATT-3′ and Reverse 5′-CGAAGAACTCCAGCATGAGA-3′. An 809 bp hph product was amplified from the hygromycin gene of pDs(Hyg)35S using forward primer 5′-GATGTAGGAGGGCGTGGATA-3′ and the reverse primer 5′-ATTTGTGTACGCCCGACAGT-3′. The C1 probe was obtained by digestion of pBECKSred with EcoRI and excision of the 1.1 kb band. All probes were gel purified using the Qiaex II kit (Qiagen, Valencia, CA).
Radioactive hybridizations were performed by denaturing the DNA probes and labeling with Easy Tides α-32P dCTP specific activity 6,000 Ci (Perkin Elmer-General Electric, Waltham, MA) using Megaprime™ random primer labeling kit (Amersham-General Electric, Piscataway, NJ). Unincorporated α-32P dCTP was removed using Nucaway™ spin columns (Ambion-Applied Biosystems, Austin, TX). Labeled probes with at least 1.0 × 106 counts per min per ml of hybridization buffer were used in hybridization. The blots were hybridized at 43–45°C using Ultrahyb™ hybridization buffer (Ambion-Applied Biosystems, Austin, TX) and washed at the recommended temperatures and stringencies. Blots were exposed to Kodak Biomax™ MS film (Fisher Scientific, Pittsburgh, PA) or exposed to BIORAD phosphorimaging screens (Hercules, CA). Phosphor images were scanned on a BIORAD phosphorimager (Hercules, CA) and the scans were analyzed using Quantity One™ software (BIORAD, Hercules, CA). Image J software (National Institute of Health; http://rsb.info.nih.gov/ij/) was used to subtract background, remove noise, and alter contrast from some of the digital images.
Reverse transcriptase PCR (RT-PCR)
RNA was extracted using the Qiagen RNeasy™ extraction kit (Valencia, CA). RNase-free DNase I polymerase was used to degrade plant genomic DNA before the RT reaction. RT-PCR was done in two steps using the Masterscript™ RT-PCR System from 5 Prime (San Francisco, CA) according to the instructions. Mastermix 1 (15 μl) was set-up as advised by the vendor. Mastermix 2 was total RNA (200 ng–1 μg in 5 μl) heated at 65°C and chilled on ice. Both tubes were mixed together and incubated for 90 min at 42°C.
PCR was performed with Taq DNA polymerase (Takara, Clonetech, Mountain View, CA) using primers: C1 Forward: 5′-TCGGACGACTGCAGC TCGGC-3′, C1 Reverse: 5′-CCTCGTGCTTATTGGACA-3′ to generate a product of 213 bp. PCR conditions were: initial denaturation at 94°C for 4 min; 30 cycles of 92°C for 30 s; 55°C for 30 s; 72°C for 30 s; and final extension at 72°C for 5 min. The NPTII primers used were NPTII Forward 5′-TGCTCCTGCCGAGAAAGTAT-3′, NPTII Reverse 5′-AATATCACGGGTAGCCAACG-3′ to generate a 356 bp product or Forward 5′-GTAGCCGGATCAAGCGTATG-3′, NPTII Reverse 5′-GCTCGACGTTGTCACTGAAG-3′ to yield a 150 bp product.
PCR conditions were: initial denaturation at 94°C for 4 min; 25 cycles of 92°C for 30 s; 60°C for 30 s; 72°C for 30 s; and final extension at 72°C for 5 min. The hph primers used were hph Forward 5′-GTGTCACGTTGCAAGACCTG-3′ and hph Reverse 5′-ACATTGTTGGAGCCGAAATC-3′ to generate a 322 bp product using the same thermal cycler program as for NPTII primers with an annealing temperature of 52°C.
Crosses with Zuzanna spring wheat
To unequivocally validate that gene transfer was nuclear, high NPTII ELISA transgenic plants were used as the pollen parents in crosses with Zuzanna, a spring wheat cultivar. Zuzanna plants were emasculated prior to pollen shed and covered in glassine bags for 2 days. Pollen from one spike of transgenic wheat line was dusted on the pistils of Zuzanna, the spikes were bagged, and allowed to set seed. Hybrid seed was collected and tested by PCR for the presence of the NPTII gene.
Results
The floral dip method has been used to produce three transformants, 3B2, 14C1 and 1CR1 in Crocus wheat and the transgenes have been characterized for stability over three to six generations. Preliminary experiments were performed by dipping plants in Agrobacterium infiltration medium 0–4 days prior to anthesis, as judged by the age of the florets in the middle of the spikes. Wheat flowering is asynchronous with the middle florets reaching anthesis prior to the terminal florets. Screening of 25,000 seedlings failed to isolate a single transformation event (J. Zale and C. Steber, unpublished). Following these negative experiments, further dipping experiments used only spikes younger than 4 days prior to anthesis. The precise stage of flowering was determined by microscopic examination of developing microspores (Table 1). 3B2 and 14C1 were produced using the C58C1 Agrobacterium strain harboring pDs(Hyg)35S. The 1CR1 transformant was produced using strain AGL1 harboring pBECKSred.
Transformation of Crocus with pDs(Hyg)35S
Two experiments using spikes at the uninucleate stage of microspore development were performed using infiltration media containing strain C58C1 carrying pDs(Hyg)35S (Table 1). The infiltration medium was formulated essentially as described for the Arabidopsis floral dip (Bechtold et al. 1993; Clough and Bent 1998) except that acetosyringone was added. Acetosyringone is a phenolic compound that induces the virulence genes in Agrobacterium (Hirooka et al. 1987). The first transformant, 3B2, was generated after dipping six unemasculated mid-late uninucleate stage spikes, with clipped florets, into infiltration media for 2 min. The spikes were bagged for 2 days to maintain humidity, and the plants were left to self-pollinate. Seed set after dipping was reduced (Table 1). The T1 seeds were sterilized with a commercial bleach solution, planted, and the seedlings sprayed with 2% paromomycin at the two to three leaf stage to screen for resistance conferred by the NPTII gene (Cheng et al. 1997). One T1 Crocus plant showed minimal spotting due to the paromomycin spray and was advanced to the T2 for further analysis. One hundred and twenty-one T2 progeny were tested for resistance to paromomycin spray. The T2 plants segregated in a 3:1 ratio of 93 resistant: 28 sensitive (χ2 = 0.187; probability (p) = not significant (NS)) suggesting one T-DNA insertion. To confirm the segregation analysis, genomic DNA was isolated from 14 T2 seedlings and analyzed in PCR with the NPTII primers. Ten out of 14 showed the presence of NPTII suggesting 3:1 segregation (χ2 = 0.095; p = NS; Fig. S2).
The second transformant, 14C1, was obtained after dipping spikes of Crocus with C58C1 harboring pDs(Hyg)35S twice for 2 min on two consecutive mornings. The wheat spikes were at the early to mid uninucleate microspore stage, when the spike was approximately four cm in length. Such double dipping is also practiced in Arabidopsis (Bent 2000). Seed set was severely reduced after the two treatments and only one T1 seed was recovered (Table 1). This T1 seedling showed minimal spotting with 2% paromomycin and was advanced to produce T2 seeds. The T2 seeds were plated on MS medium with hygromycin (60 mg/L) to determine segregation ratios. Segregation in the T2 generation produced 275 hygromycin resistant:25 hygromycin sensitive plants (15:1; χ2 = 2.02, p = NS) suggesting the presence of two T-DNA insertions. Sensitive plants were very chlorotic.
Southern analysis was used to verify the transformation of 3B2 and 14C1 progeny and to show transgene transmission over several generations. Left border analysis was used to determine the number of transgenes inserted into the genome (Cheng et al. 1997). Plant genomic DNA was digested with EcoR1 and genomic DNA blots were hybridized with an NPTII probe (Fig. 1a). The single EcoR1 site within the pDs(Hyg)35S T-DNA is to the right of the probe; the next EcoR1 site to the left of the NPTII probe is upstream within the wheat genomic DNA. The NPTII probe showed a single 3.0 kb band in 3B2 indicating a single insertion. This 3.0 kb band was observed by Southern analysis in the T4 (Fig. S3a), the T5 (Fig. S3b), and in the T6 generations (Fig. 3a). Identification of the same band in multiple generations indicates stable transmission of the transgene. The same left border analysis identified a 2.6 kb and a 4.0 kb band in the T2 progeny of 14C1 indicating two insertion events (Fig. 3a). This is consistent with the fact that hygromycin resistance segregated 15:1 in the T2 generation.
Molecular analyses of pDs(Hyg)35S transformants. a Southern hybridization of DNA gel blots with the NPTII probe showing copy number of T6 progeny of 3B2 and T2 progeny of 14C1. Genomic DNA of WT Crocus, 3B2 and 14C1 were digested with EcoRI and probed with the NPTII probe. b BglII digested genomic DNA was blotted and probed with the Hph probe in T6 progeny of 3B2, T5 progeny of 14C1, and WT Crocus. Ethidium bromide stained genomic DNA loading controls are shown at the top of each blot. c PCR showing the transmission of the NPTII transgene transmission to Zuzanna/14C1 F1 hybrids. 14C1 and Zuzanna are included as positive and negative controls
A different Southern analysis strategy was used to analyze the hygromycin markers in 14C1 and 3B2. Genomic DNA was digested with BglII, an enzyme that cuts on either side of the hygromycin transgene and probe in the pDs(Hyg)35S T-DNA (Fig. 1a). The presence of the hygromycin transgene results in an 800 bp band when a DNA blot is hybridized with the hygromycin probe in T6 progeny 3B2, and in the T5 progeny of 14C1, thus confirming transmission of the hygromycin transgene (Fig. 3b).
Finally, to demonstrate unequivocally that the T-DNA had integrated into the nuclear genome of the transgenic plants, 14C1 T2 transformants were used as the pollen parents in crosses with ‘Zuzanna’ wheat as the female parent. Two F1 ‘Zuzanna’/14C1 hybrid seeds were germinated and the seedlings were tested for the presence of the NPTII transgene by PCR. The ‘Zuzanna’ parent tested negative, whereas the F1 hybrids tested positive demonstrating that the NPTII transgene is transmitted in crosses via the pollen (Fig. 3c).
Transformation of Crocus with pBECKSred
In working with the pDs(Hyg)35S transformants, one limitation in identifying transformants was the need to use the paromomycin screen to identify primary transformants. Thus, additional floral dip experiments were performed using pBECKSred, a T-DNA vector containing both the NPTII marker and two maize regulators of anthocyanin synthesis, C1 and Lc, whose expression allowed a visual screen for red pigmentation in transformed seed.
Floral dip experiments with infiltration media containing Agrobacterium AGL1 harboring pBECKSred (McCormac et al. 1997) were performed on Crocus spikes at different stages of microspore development including the late uninucleate, the binucleate, and trinucleate stages (Table 1). No transformation events, as determined by red embryos, screens for paromomycin resistance and NPTII ELISAs, were isolated in 3,000 T1 seeds derived from dipping of binucleate stage spikes or in 4,200 T1 seeds derived from dipping of trinucealate stage spikes (Table 1). It was observed that a small number of seeds from these experiments did show red foci on the endosperm; however, red pigmentation on the endosperm was not sufficient to identify transformants.
Earlier stages of treatment were tested in dipping experiments, and six spikes of Crocus at the late uninucleate microspore stage were dipped for two minutes in infiltration media. The spikes were covered in plastic bags for 2 days and allowed to self-pollinate. Seed set was not reduced to a great extent presumably because the spikes were older (Table 1). Out of 33 T1 seeds screened, 1CR1 and 1CR2, had dark red embryos and were shriveled relative to wild-type (Fig. S1). These red seeds were germinated and T1 seedlings were screened with paromomycin and the NPTII ELISA assays. Only one of the putative transformants, 1CR1, was positive. Southern analysis was used to detect the presence of the NPTII gene by digesting T1 genomic DNA with two enzymes that cut on either side of the NPTII transgene and probe, EagI and EcoRI. Only 1CR1 showed the expected 1,208 bp band (Fig. 1b) indicating the presence of the NPTII transgene (Fig. 4). The 1CR1 plant was self-pollinated to produce T2 seed.
Southern analyses of 1CR1. a Eag1 and EcoRI digested DNA gel blot of 1CR1, 1CR2, and WT Crocus probed with NPTII detects a 1.2 kb band internal to the T-DNA insert. b Left border analysis for copy number was performed using the NPTII probe and BamHI digested genomic DNA of 1CR1 T2 and WT Crocus. EcoR1-digested 3B2 was included as a positive control. The single 5.5-kb band indicates a single insertion event in 1CR1. c HindIII digested DNA gel blot of WT Crocus and 1CR1 T3 probed with the C1. Ethidium bromide stained DNA loading controls are shown on top of each blot
Southern analysis was used to show integration of the T-DNA transgene into the wheat genome, to determine transgene copy number, and to detect transgene transmission to the T2 and T3 generation. Left border analysis was performed by digesting T2 and T3 genomic DNA with BamHI, an enzyme that cuts to the right of the NPTII probe (Fig. 1b). Hybridization of genomic DNA blots with the NPTII probe detected a single band of 5.5 kb in 1CR1 T2 (Fig. S4) and T3 plants (Fig. 4b). This indicates that a single copy of the NPTII transgene was stably transmitted over the T2 and T3 generations.
If the T-DNA of pBECKSred is integrated into the Crocus genome, the 2.5 kb C1 transgene should also be present. 1CR1 T3 genomic DNA was digested with HindIII, an enzyme cuts on either side of the C1 probe resulting in a band of ~2.5 kb (Fig. 1). Hybridization of DNA blots with the C1 probe detected the expected 2.5 kb C1 band in 1CR1, but not in Crocus wild-type (Fig. 4c). This confirmed the transformation of 1CR1 with the pBECKSred T-DNA.
Segregation analysis of T2 seeds was performed by examining T2 seeds under the stereoscope and assessing differential endosperm seed coat color, presumably due to segregation of progeny expressing the Lc/C1 genes. Sixty-seven out of 96 T2 seeds displayed an enhanced red seed coat color, suggesting a 3:1 segregation of the transgene (χ2 = 1.41; p = NS). An example of one seed with red seed endosperm coat color compared to the wild-type Crocus is shown in Fig. 5. To validate the segregation result based on seed color, a random population of T2 seedlings (N-31) was scored for paromomycin resistance. This yielded a segregation ratio of tolerant:sensitive of 21:10 (3:1; χ2 = 0.871; p = NS) which is consistent with Southern analysis data indicating a single insertion of the transgene. From 13 paromomycin progeny, triplicate extracts were analyzed for the presence of the NPTII protein using an ELISA assay (Table S1). A total of nine were positive for NPTII by ELISA assay.
Reverse transcriptase (RT)-PCR
RT-PCR was performed on progeny derived from 3B2, 14C1 and 1CR1 in order to examine transgene expression. For the two pDs(Hyg)35S transformants, NPTII expression was detected in the T6 progeny of 3B2 and the T5 progeny of 14C1 (Fig. 6a). Expression of the hygromycin resistance gene was also detected (Fig. 6b). Finally, RT-PCR was performed using C1 primers to detect C1 mRNA expression, driven by the 5S:adh1 intron promoter, in T3 progeny of 1CR1 (Fig. 6c).
Discussion
This research establishes that the floral dip can be used to isolate stable low-copy number transformants in wheat without the use of tissue culture. Three well-characterized transformants have been isolated in the germplasm line Crocus, and Southern analysis has shown the transgenes to be integrated in the wheat genome and are stable for three–six generations. The transgenes showed the expected segregation ratios for a nuclear insertion event, and this was verified by transmission through pollination. Transgene expression was observed in the T5 and T6 generations.
The developmental stage of the flowers dipped proved to be crucial for successful floral transformation of wheat. In the early attempts at wheat transformation, Agrobacterium infiltration medium harboring various genetic constructs with acetosyringone and Tween 20 was pipetted into uncut florets of wheat at or near the time of anthesis in an effort to transform the pollen (Hess et al. 1990; Langridge et al. 1992). Langridge et al. (1992) failed to demonstrate transmission of the transgene to the next generation. It is possible that such transformation events obtained late in floral development, if obtained at all, may have a tendency toward generating chimeras. In the present study, preliminary research failed to identify a single transformation event in 25,000 putative transformants produced by dipping at stages closer to anthesis (i.e. within 4 days prior to anthesis; J. Zale and C.M. Steber, unpublished). Similarly, no transformants were produced in Arabidopsis if dipping occurred later than 4 days before anthesis (Clough and Bent 1998). Conversely, in wheat, little or no seed set was obtained when florets are dipped too early when the spike was approximately 4 cm long. The ideal stage for dipping appears to be the mid to late uninucleate microspore stage when the spike has not emerged from the sheath. The spike will be approximately 6–7 cm long at this stage and, depending on environmental conditions, approximately 4–7 days before anthesis. This stage coincides with that preferentially chosen for microspore culture (Liu et al. 2002). The target tissue for the T-DNA transfer to wheat has not been determined, but based on the work in Arabidopsis, the ovule is the likely target for T-DNA transfer (Bechtold et al. 2000; Desfeux et al. 2000; Ye et al. 1999).
The transformation efficiency in these experiments was estimated at 0.44% (number of transformants/number of florets dipped). The originally reported transformation efficiency of Arabidopsis by the floral dip ranged between 0.23 and 0.47% in the presence of 0.005% Silwet L-77 (Clough and Bent 1998). If transformation efficiency is reported as the number of transformants/the number of seeds set, then the efficiency is much higher at 6.8% for these three experiments. The latter transformation efficiency was relatively high due to low seed set in the production of 14C1. It appears that dipping at the early uninucleate microspore stage, and/or dipping twice, in an Agrobacterium infiltration medium containing Silwet L-77 significantly decreases seed set in wheat (Table 1). Similarly, any other stress event such as disease, heat, drought, or covering the spikes with heavy glassine bags lowers seed set.
One of the most important considerations in identifying transformed plants is the choice of a selectable marker or reporter genes that are reliable indicators of the transformation event. The maize Lc/C1 anthocyanin regulators in pBECKSred (McCormac et al. 1997) turn tissue red and can be used to identify transformation events by visually scoring for red embryos in T1 seed. However, the stress of dipping can also produce false positives and induce anthocyanin accumulation in the seed, particularly in a red wheat variety. Therefore, using a secondary screen would be appropriate.
The choice and concentration of selection agent are also important considerations when using this protocol. Wheat has a naturally high tolerance to many antibiotics when seeds are plated on nutrient medium (Langridge et al. 1992) and in hydroponics (D. J. Guerra and C. M. Steber, unpublished). However, the concentration of selection agent should not be so high as to be lethal to low copy number T-DNA transformants. We chose not to use geneticin G418 or any other kanamycin analog in nutrient medium because of the prevalence of escapes with NPTII selection (Rakosy-Tican et al. 2007). Screening based on the bleaching induced by the 2% paromomycin spray (Cheng et al. 1997) is, at times, subjective and influenced by environmental conditions. Under higher temperatures, plant tissue will become desiccated and this may confound scoring. One must also consider the strength of the promoter when assessing tolerance or sensitivity and compare the plants tested against wild-type control plants that have also been sprayed.
The large size of the wheat genome at 17,000 Mbp per haploid nucleus (Bennett and Leitch 1996) makes it difficult to detect low copy number T-DNA transfer to wheat. Dirty genomic DNA preparations, degraded DNA, problems with DNA transfer to the membrane, and poor probe labeling can yield negative results in Southern analysis. Conversely, related sequences may be detected due to homology with the probe if the washes are not sufficiently stringent. Langridge et al. (1992), detected bands at 800 bp and 8 kb band on EcoRI and EcoRI/PstI digested Southern blots with the NPTII probe in the T0 generation after treating wheat, barley and maize plants with Agrobacterium, but these bands were not transmitted to the next generation. The authors concluded that these bands might have been due to transformation of an endophytic bacterium. The present study occasionally detected faint 500–800 bp bands in advanced progeny of 3B2 when EcoRI-digested Southern blots were hybridized with the NPTII probe. This band may be due to incomplete transfer of the NPTII transgene at the left border.
Three well-characterized transformations events isolated in the ‘Crocus’ genotype show that the method is reproducible and can produce transformation events that are stable over three–six generations. Transformants have also been produced in Chinese Spring and Bobwhite, however further work is needed to determine whether some genotypes are more amenable than others. A significant obstacle to obtaining transgenic wheat seed using this method is the low seed following dipping. Future work will optimize this protocol so that transformants can consistently be produced while maximizing seed set. These experiments will test different Agrobacterium strains, alternative selectable markers, and different surfactants.
In conclusion, we have produced low copy number T-DNA transformants in Crocus wheat using a floral transformation protocol. The transgenes have integrated into the plant genome, are stable over several generations as determined by Southern analysis, are transmitted in crosses, and are expressed in progeny derived from the primary transformants.
Abbreviations
- NPTII :
-
Neomycin phosphotransferase gene
- Lc/C1 :
-
Anthocynanin regulatory genes of maize
- Hph :
-
Hygromycin gene
- Ds :
-
Dissociation element of maize
- PCR:
-
Polymerase chain reaction
- RT-PCR:
-
Reverse transcriptase-PCR
- ELISA:
-
Enzyme-linked immunosorbent assay
- MS:
-
Murashige–Skoog
- LB:
-
Left border
- RB:
-
Right border
References
An G, Lee S, Kim SH, Kim SR (2005) Molecular genetics using T-DNA in rice Plant. Cell Physiol 46:14–22
Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci 316:1194–1199
Bechtold N, Jaudeau B, Jolivet S, Maba B, Vezon D, Voisin R, Pelletier G (2000) The maternal chromosome set is the target of the T-DNA in the in planta transformation of Arabidopsis thaliana. Genetics 155:1875–1887
Bennett MD, Leitch IJ (1996) Nuclear DNA amounts in Angiosperms. Ann Bot 76:113–176
Bent AF (2000) Arabidopsis in planta transformation Uses, mechanisms, and prospects for transformation of other species. Plant Physiol 124:1540–1547
Cheng M, Fry JE, Pang S, Zhou H, Hironaka CM, Dr Duncan, Conner TW, Wan Y (1997) Genetic transformation of wheat mediated by Agrobacterium tumefaciens. Plant Physiol 115:971–980
Chomczynski P (1993) One-hour downward alkaline capillary transfer for blotting of DNA and RNA. Anal Biochem 201:134–139
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Desfeux C, Clough SJ, Bent AF (2000) Female reproductive tissues are the primary target of Agrobacterium-mediated transformation by the Arabidopsis floral-dip method. Plant Physiol 123:895–904
Hess D, Dressler K, Nimmrichter R (1990) Transformation experiments by pipetting Agrobacterium into the spikelets of wheat (Triticum aestivum L.). Plant Sci 72:233–244
Hirooka T, Rogowsky PM, Kado CI (1987) Characterization of the virE locus of Agrobacterium tumefaciens plasmid pTiC58. J Bacteriol 169:1529–1536
Jones HD (2005) Wheat transformation: current technology and applications to grain development and composition. J Cereal Sci 41:137–147
Jordan MC (2000) Green fluorescent protein as a visual marker for wheat transformation. Plant Cell Rep 19:1069–1075
Kaeppler SM, Kaeppler HF, Rhee Y (2000) Epigenetic aspects of somaclonal variation in plants. Plant Mol Biol 43:179–188
Kleinhofs A, Kilian A, Saghai-Maroof MA, Biyashew RM, Hayes P, Chen FQ, Lapitan N, Fenwick A, Blake TK, Kanazin V, Ananiev E, Dahleen L, Kudrna D, Bollinger J, Kanpp SJ, Liu B, Sorrells M, Heun M, Franckowiak JD, Hoffman D, Scadsen R, Steffensen BJ (1993) A molecular, isozyme, and morphological map of the barley (Hordeum vulgare) genome. Theor Appl Genet 86:705–712
Langridge P, Brettschneider R, Lazzeri P, Lorz H (1992) Transformation of cereals via Agrobacterium and the pollen pathway: a critical assessment. Plant J 2:631–638
Lazo GR, Stein PA, Ludwig RA (1991) A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology (N Y) 9:963–967
Liu W, Zheng MY, Polle EA, Konzak CF (2002) Highly efficient doubled-haploid production in wheat (Triticum aestivum L.) via induced microspore embryogenesis. Crop Sci 42:686
Long D, Goodrich J, Wilson K, Sundberg E, Martin M, Puangsomlee P, Coupland G (1997) Ds elements on all five Arabidopsis chromosomes and assessment of their utility for transposon tagging. Plant J 11:145–148
McCormac AC, Wu H, Bao M, Wang YQ, Xu R, Elliot MC, Chen DF (1997) The use of visual marker genes as cell-specific reporters of Agrobacterium-mediated T-DNA delivery to wheat and barley. Euphytica 99:17–25
Mead R, Curnow RN, Hasted AM (2003) Statistical methods in agriculture and experimental biology. Chapman and Hall/CRC, Boca Raton
Mohan Jain S (2001) Tissue culture-derived variation in crop improvement. Euphytica 118:153–166
Murashige T, Skoog FA (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15:473–497
Nehra N, Chibbar R, Leung N, Caswell K, Mallard C, Steinhauer L, Baga M, Kartha K (1994) Self-fertile transgenic wheat plants regenerated from isolated scutellar tissues following microprojectile bombardment with two distinct gene constructs. Plant J 5:285–297
Pacurar DI, Thordal-Christensen H, Nielsen KK, Lenk I (2007) A high-throughput Agrobacterium-mediated transformation system for the grass model species Brachypodium distachyon L. Transgenic Res 17:965–975
Pellegrineschi A, Noguera LM, Skovmand B, Brito RM, Velazquez L, Salagdo MM, Hernandez R, Warburton M, Hoisington D (2002) Identification of highly transformable wheat genotypes for mass production of self-fertile plants. Genome 45:420–421
Rakosy-Tican E, Aurori CM, Dijkstra C, Thieme R, Aurori A, Davey MR (2007) The usefulness of the gfp reporter gene for monitoring Agrobacterium-mediated transformation of potato dihaploid and tetraploid genotypes. Plant Cell Rep 26:661–671
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Supartana P, Shimizu T, Nogawa M, Shioiri H, Nakajima T, Haramoto N, Nozue M, Kojima M (2006) Development of simple and efficient in planta transformation method for wheat (Triticum aestivum L.) using Agrobacterium tumefaciens. J Biosci Bioeng 102:162–170
Taylor NJ, Fauquet CM (2002) Microparticle bombardment as a tool in plant science and agricultural biotechnology. DNA Cell Biol 21:963–977
Vasil V, Castillo A, Fromm M, Vasil IK (1992) Herbicide resistance fertile transgenic wheat plants obtained by micro-projectile bombardment of regenerable embryogenic callus. Biotechnology 1:667–674
Weeks JT, Anderson OD, Blechl AE (1993) Rapid production of multiple independent lines of fertile transgenic wheat (Triticum aestivum). Plant Physiol 102:1077–1084
Wilson K, Long D, Swinburne J, Coupland G (1996) A dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell 8:659–671
Wu H, Doherty A, Jones HD (2007) Efficient and rapid Agrobacterium-mediated genetic transformation of durum wheat (Triticum turgidum L. var. durum) using additional virulence genes. Transgenic Res 17:425–4356
Xiang-Yuan X, De-Cai C (1983) Relationship between pollen and embryo sac development in wheat, Triticum aestivum L. Bot Gaz 144:191–200
Ye GN, Stone D, Pang SZ, Creely W, Gonzalez K, Hinchee M (1999) Arabidopsis ovule is the target for Agrobacterium in planta vacuum infiltration transformation. Plant J 19:249–257
Zale J, Scoles G (1999) Registration of Crocus hard red spring wheat. Crop Sci 39:1539–1540
Zale J, Borchardt-Wier H, Kidwell K, Steber C (2004) Callus induction and plant regeneration from mature seed embryos of a diverse set of wheat genotypes. Plant Cell Tissue Organ Cult 76
Zhao TJ, Zhao SY, Chen HM, Zhao QZ, Hu ZM, Hou BK, Xia GM (2006) Transgenic wheat progeny resistant to powdery mildew generated by Agrobacterium inoculum to the basal portion of wheat seedling. Plant Cell Rep 25:1199–1204
Acknowledgments
This material is based upon work supported by the National Science Foundation under Grant No. 0638421 (to J.Z.) and the USDA NRI Grant No. 2001-01856 (to J.Z. and C.M.S.). We gratefully acknowledge Dr. L. Ohnoutkova for determining the stage of microspore development and for providing seed of Zuzanna spring wheat.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. Jones.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Zale, J.M., Agarwal, S., Loar, S. et al. Evidence for stable transformation of wheat by floral dip in Agrobacterium tumefaciens . Plant Cell Rep 28, 903–913 (2009). https://doi.org/10.1007/s00299-009-0696-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-009-0696-0