Abstract
In eukaryotes, all DNA transactions happen in the context of chromatin that often takes part in regulatory mechanisms. In particular, chromatin structure can regulate exchanges of DNA occurring through homologous recombination. Few systems have provided as detailed a view on this phenomenon as mating-type switching in yeast. Mating-type switching entails the choice of a template for the gene conversions of the expressed mating-type locus. In the fission yeast Schizosaccharomyces pombe, correct template choice requires two competing small recombination enhancers, SRE2 and SRE3, that function in the context of heterochromatin. These two enhancers act with the Swi2/Swi5 recombination accessory complex to initiate strand exchange in a cell-type-specific manner, from SRE2 in M cells and SRE3 in P cells. New research indicates that the Set1C complex, responsible for H3K4 methylation, and the Brl2 ubiquitin ligase, that catalyzes H2BK119 ubiquitylation, participate in the cell-type-specific selection of SRE2 or SRE3. Here, we review these findings, compare donor preference in S. pombe to the distantly related budding yeast Saccharomyces cerevisiae, and contrast the positive effects of heterochromatin on the donor selection process with other situations, where heterochromatin represses recombination.
Similar content being viewed by others
Introduction
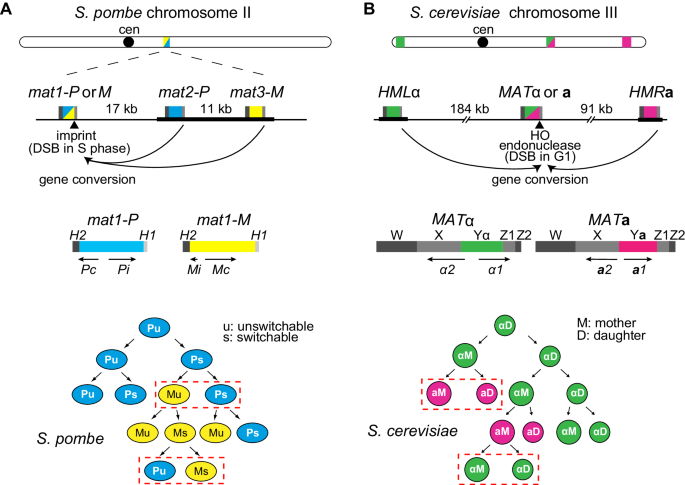
The fission yeast S. pombe and the budding yeast S. cerevisiae both switch mating type during vegetative growth (Fig. 1). Schizosaccharomyces pombe switches between the P and M cell types by replacing the content of the expressed mat1 cassette with information copied from one of the two linked silent cassettes, mat2-P or mat3-M (reviewed by Klar et al. 2014). Saccharomyces cerevisiae switches between the a and α cell types by replacing the content of the MAT locus located on chromosome III with information copied from the silent HMLα or HMRa donors located at either end of the same chromosome (reviewed by Lee and Haber 2015). In both yeasts, mating-type switching is initiated by a double-strand break (DSB), which is repaired by ectopic homologous recombination (gene conversion); but the origin of the DSB is quite different in the two distantly related yeasts. In fission yeast, a single-strand nick or gap is “imprinted” at mat1 in the S phase of the cell cycle, so that the following round of DNA replication creates a DSB on one of the two sister chromatids (Arcangioli and de Lahondes 2000; Dalgaard and Klar 2001). In budding yeast, a site-specific HO endonuclease creates the break at MAT (Kostriken et al. 1983; Strathern et al. 1982), normally at the end of the G1 phase (Nasmyth 1983). In both yeasts, mating-type switching depends on the key homologous recombination protein Rad51 and on its principal mediator Rad52 (Aboussekhra et al. 1992; Malone and Esposito 1980; Ostermann et al. 1993; Roseaulin et al. 2008). Briefly, a Rad51-coated end of the DSB promotes strand invasion into a short region of homology shared by the mat1/MAT locus and its donors (H1 in S. pombe, Z in S. cerevisiae), followed by copying of the mating-type-specific P/M or Ya/Yα regions and terminating in the H2 or W regions of homology shared on the opposite side of the DSB (Fig. 1).
Mating-type cassettes and mating-type switching patterns in S. pombe (a) and S. cerevisiae (b). In S. pombe, the expressed mat1 locus switches between mat1-P and mat1-M due to gene conversions by the linked mat2-P and mat3-M silent loci. In S. cerevisiae, the expressed MAT locus switches between MATα and MATa due to conversions by HMLα and HMRa. Silent heterochromatic regions are indicated by thick black lines. Gene conversions are initiated in both cases by a DNA break at the expressed locus followed by strand invasion at one of the silent cassettes. Strand invasion occurs at homology boxes present at the three cassettes: H1 in S. pombe and Z1 in S. cerevisiae. In S. pombe, the DNA break is formed during DNA replication in cells that have acquired an imprint during the previous DNA replication (switchable cells), and only one of the newly synthesized chromatids undergoes conversion producing one switched and one unswitched progeny (examples framed in red in cell pedigree). In S. cerevisiae, the DNA break is formed and used in the G1 phase of the cell cycle, exclusively in mother cells due to restricted expression of the HO endonuclease in mother cells, this produces pairs of switched cells after DNA replication and cell division (examples framed in red). The silent cassettes that contain mating-type information opposite to the expressed cell type are preferentially chosen in both yeasts, giving rise to the observed high efficiency of switching in the progeny of switching-competent cells; however, donor preference is not determined by the mating-type-specific sequences in the donors
The precise molecular mechanisms of switching differ greatly between the two yeasts, but they have in common their remarkable efficiency, and the fact that in both cases heterochromatic templates are used as donors for the gene conversions of the expressed cassette. Thus, while observations in many organisms have indicated that recombination can be inhibited in heterochromatic regions to limit exchanges between repetitive elements, the mating-type systems provide a more nuanced view of the effects of chromatin structure on recombination by showing how various histone modifications, in particular heterochromatic modifications in fission yeast, can instead promote the choice of gene conversion donors and efficient recombination.
Chromatin factors required for directional mating-type switching in fission yeast
Three genetic screens have produced S. pombe mutants with donor choice defects (Egel et al. 1984; Thon et al. 2005; Maki et al. 2018). Donor choice, or directionality of mating-type switching, refers to a selection process, whereby the mat2-P cassette is preferentially chosen in M cells to convert the expressed mat1 locus, while the mat3-M cassette is preferentially chosen in P cells, resulting in equal proportions of the two cell types (Fig. 1) (Egel and Eie 1987; Klar 1990; Miyata and Miyata 1981). Heterochromatin defects are at the source of impaired donor choice for many mutants. Thus, a mutant in the chromodomain protein Swi6, homolog of the major heterochromatin protein HP1 in other organisms, was obtained in the first genetic screen for mating-type switching deficient mutants (Egel et al. 1984) and subsequently characterized for its directionality defects (Jakociunas et al. 2013; Jia et al. 2004; Thon and Klar 1993). Another strategy identified the multi-subunit Clr4-containing complex (CLRC) responsible for the methylation of histone H3K9 as necessary for proper donor choice, by searching for mutations that reduce heterologous switching in the wild-type h90 strain but increase heterologous switching in the h09 strain, where the mat2 and mat3 cassette contents are swapped (Thon et al. 2005). More recently, we used a sensitive high-throughput fluorescence microscopy approach to find mutants with aberrant ratios of P and M cells, among which directionality mutants were identified (Maki et al. 2018). Table 1 presents the mutants obtained in this screen that have defects in histone-modifying enzymes. The list emphasizes that accurate donor selection requires the H3K9 modification pathway and various histone deacetylases (HDACs), as well as modifications whose relevance to switching had previously not been suspected. In particular, the screen identified the Set1C complex responsible for histone H3K4 methylation and the ubiquitin ligase Brl2 responsible for histone H2BK119 ubiquitylation. Mutations in these factors lead to less extreme biases in mating-type distributions than mutations in heterochromatin factors.
Heterochromatin normally occupies ~ 20 kb in the mating-type region including the mat2-P and mat3-M cassettes (Fig. 2). It is formed by the histone H3K9 methyltransferase CLRC and by several HDACs including the NAD-dependent Sir2, the Class I HDAC Clr6, and the SHREC complex containing the Class II HDAC Clr3. Heterochromatinization culminates in the recruitment of two HP1 homologs, Swi6 and the chromodomain protein Chp2. An important function of heterochromatin is to silence mat2-P and mat3-M, but due to redundant mechanisms of silencing, a strong repression remains in CLRC, Swi6, and Chp2 mutants, while a small derepression is seen in SHREC mutants (Hansen et al. 2011; Maki et al. 2018; Thon and Verhein-Hansen 2000). A derepression in the same order occurs in swi6Δ and chp2Δ mutants, yet chp2Δ mutants do not display switching defects (Thon and Verhein-Hansen 2000), showing that directionality defects in heterochromatin mutants are not a mere consequence of deregulated cell-type expression. Rather, heterochromatin regulates the use of two competing recombination enhancers, SRE2 and SRE3 (see below).
Heterochromatin at the donor loci. a In S. pombe, histone deacetylation by several HDACs (SHREC, Clr6, Sir2) together with histone H3K9 methylation by CLRC forms heterochromatin over a 20 kb domain between the IR-L and IR-R boundaries. Following histone modification, the chromodomain protein Swi6 associates with the entire 20 kb region. cenH is an RNA-interference heterochromatin nucleation center with centromere homology. b In S. cerevisiae, heterochromatin results from histone deacetylation and association of Sir proteins in two smaller domains, each < 3 kb, at the HML and HMR loci. At each cassette, the E and I silencers recruit various combinations of DNA-binding proteins, ORC, Rap1, and Abf1, to initiate heterochromatin formation
A novel aspect revealed by our recent screen is that the directionality of mating-type switching also relies on Set1C and its partner, the Brl2 ubiquitin ligase (Maki et al. 2018). Individual deletions of six of the eight Set1C subunits alter donor choice (Table 1). The identified mutations severely impair methylation of histone H3K4, unlike mutations in the two subunits that were not identified in the screen, Shg1 and Sdc1 (Mikheyeva et al. 2014). Furthermore, a necessary step for H3K4 methylation is ubiquitylation of histone H2BK119; both modifications fail in the Brl2 mutant (Zofall and Grewal 2007). Thus, collectively, the obtained mutants points to histone H3K4 as a methylation substrate relevant to directionality and indicate that Set1C and Brl2 might contribute together with heterochromatin to the regulation of SRE2 and SRE3.
The cell-type-specific recombination enhancers SRE2 and SRE3
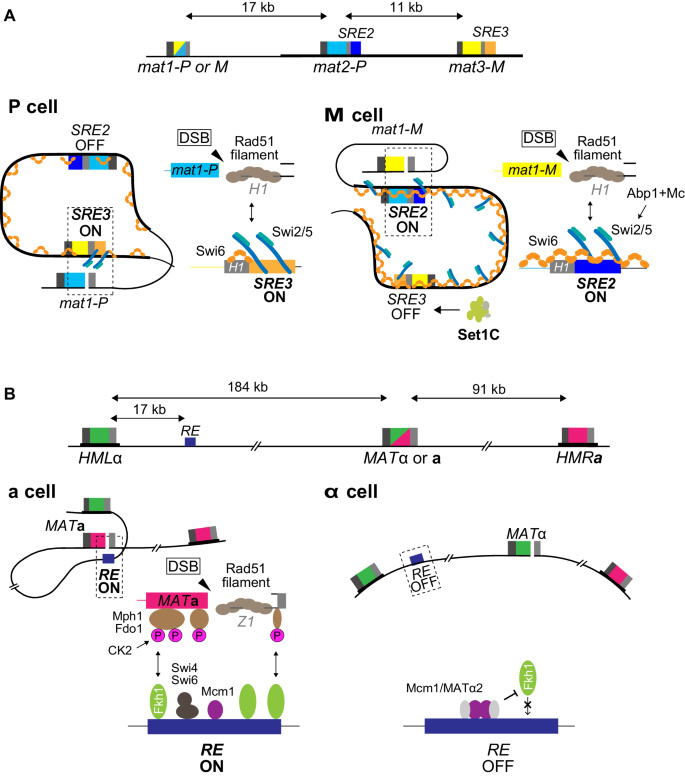
The small Swi2- dependent recombination enhancers, SRE2 and SRE3, are necessary for the cell-type-specific choice of mat2-P in M cells and mat3-M in P cells (Jakociunas et al. 2013; Jia et al. 2004) (Fig. 3). The molecular mechanism of action and regulation of these elements are not precisely known. During DNA replication, a single-ended double-strand break formed at the mat1 locus by the incoming fork initiates a template switch for leading-strand synthesis (Arcangioli and de Lahondes 2000; Dalgaard and Klar 2001). The broken molecule invades one of the silent cassettes at a 59 bp region of sequence homology, the H1 box. SRE2 and SRE3 are adjacent to the H1 boxes at mat2-P and mat3-M, respectively; each element is < 500 bp. In all likelihood, SRE2 and SRE3 direct donor choice by favoring strand invasion at their adjacent H1 box. Deletion of one enhancer results in the near exclusive use of the other cassette, whereas substitution of one enhancer with the other results in relatively efficient heterologous switching, indicating that each enhancer can stimulate recombination effectively in both P and M cells and that the two enhancers compete with each other when both are present (Jakociunas et al. 2013). Their relative use would be biased by differences in chromatin structure and differential association of the switching factor Swi2 with the mating-type region in the two cell types to achieve directionality.
Choice of recombination enhancers in S. pombe and S. cerevisiae. aSchizosaccharomyces pombe SRE2 and SRE3 enhancers. In P cells, the Swi2/Swi5 complex is exclusively associated with the recombination enhancer SRE3; this facilitates Rad51-mediated strand invasion at the H1 region of mat3 and thus promotes switching from mat1-P to mat1-M. In M cells, the Swi2/Swi5 complex is more broadly associated with the entire heterochromatic domain, this association requires the chromodomain protein Swi6. Under these conditions, the SRE2 enhancer is preferred over SRE3 leading to switching from mat1-M to mat1-P. The use of SRE3 in M cells might be limited by Set1C. bSaccharomyces cerevisiae RE enhancer. In MATa cells, RE, bound by multiple copies of the Fkh1 protein, establishes a physical contact with the MAT locus through the interaction of Fkh1’s phospho-threonine binding domain with phosphorylated proteins located at or near the DSB; these proteins include Mph1 and Fdo1. RE binding at the site of the DSB shortens the distance between MATa and HMLα, favoring Rad51-mediated recombination between MAT and HMLα. In MATα cells, binding of Fkh1 to RE is disrupted by the repressor, Mcm1/Matα2; consequently, HMRa, closer to MAT, is then preferred
Chromatin-driven association of recombination factors with SRE2 and SRE3
The preponderance of evidence indicates that Swi2 has a role in strand exchange, similar to the related Sfr1 protein (Swi five-dependent recombination repair protein 1) for which a role in Rad51-mediated strand exchange has been extensively documented (Akamatsu et al. 2003, 2007; Argunhan et al. 2017; Haruta, et al. 2006; Ito et al. 2018; Kurokawa et al. 2008). Swi2 and Sfr1 each engage the recombination accessory factor Swi5 to form respectively the Swi2/Swi5 and Sfr1/Swi5 complexes that physically and functionally interact with the Rad51 recombinase. Another key physical interaction of Swi2, with strong implications for mating-type switching, is with the chromodomain protein Swi6 (Akamatsu et al. 2003; Jia et al. 2004). Through this interaction, Swi2 bridges heterochromatin to the recombination apparatus (Fig. 3). Sfr1 on the other hand fails to interact with Swi6 and has no obvious role in mating-type switching (Akamatsu et al. 2003), while Rad51 is essential for the repair of the break at mat1 and thus the viability of switching-competent cells (Roseaulin et al. 2008).
A role for the Swi2/Swi5 complex in directionality is indicated by the requirement for both factors for efficient mating-type switching (Egel et al. 1984) and by the differential association of the complex with the mating-type region in P and M cells, in a manner that precedes break formation at mat1 (Fig. 3) (Jia et al. 2004). In P cells, the complex is predominantly at SRE3, perhaps through direct binding of Swi2 to SRE3 DNA via the Swi2 AT-hook motifs. The complex shows a broader distribution in M cells, where it covers the whole mat2-mat3 region including both SRE2 and SRE3. Unlike for its localized association with SRE3, the extended association of Swi2/Swi5 in M cells requires Swi6 (Jia et al. 2004). During mating-type switching, interaction between the Rad51 presynaptic filament originating from mat1 with Swi2/Swi5 bound to one of the SRE elements would stimulate the strand-exchange activity of the recombinase.
How differential Swi2 recruitment or activity might occur and lead to differential use of SRE2 and SRE3 in the two cell types and how other factors might influence use of SRE2 and SRE3 remain open questions. It has been proposed that two Swi2 isoforms exist, a shorter form of the protein would be produced exclusively in M cells following transcription initiation of the swi2 gene at an internal promoter under control of the Abp1 protein and the mat1-M product Mc, and this smaller form would be necessary for the broad distribution of Swi2 over the mating-type region in M cells (Yu et al. 2012) (Fig. 3). The two Swi2 isoforms have not been detected at the protein level (Jia et al. 2004; Matsuda et al. 2011), but further investigations might reveal them. Other differences between P and M cells are that the Swi2 and Swi5 proteins are in greater abundance in M cells (Matsuda et al. 2011) and more Swi6 is detected at the mating-type region in M cells (Noma et al. 2001), both of which might contribute to the increased and broader association of Swi2 and Swi5 observed in M cells and to the differential use of SRE2 and SRE3 in the two cell types.
Possible points of action for Brl2 and Set1C
A central question relates to the selective use of the SRE2 recombination enhancer in M cells, where the Swi2/Swi5 complex is detected at both SRE2 and SRE3. Greater proximity of the mat2 donor to mat1 in the linear DNA was first suggested as a determinant factor (Jia et al. 2004), yet linear proximity might not be crucial, because mat2 is still preferred when mat2 and its SRE2 enhancer are moved away from mat1 by swapping the mat2 and mat3 donor cassettes together with their respective enhancers (Jakociunas et al. 2013). Directionality still operates in these cells. One possibility is that the chromatin structure specific to M cells allows SRE2 to outcompete SRE3. Our new study (Maki et al. 2018) indicates that Set1C and Brl2 might be part of this mechanism by inhibiting SRE3. Cells whose SRE2 element is deleted use SRE3, leading to a strong bias towards M cells in the cell population (Jakociunas et al. 2013). This bias is even more pronounced in the set1Δ and brl2Δ mutants indicating Set1 and Brl2 normally limit recombination at SRE3 (Maki et al. 2018). At another locus, the ste11 gene promoter, H2BK119 ubiquitylation and H3K4 methylation prevent chromatin remodeling by the RSC complex (Materne et al. 2015, Materne et al. 2016), suggesting remodeling might also modulate recombination in the mating-type region.
Other effects are not excluded. As for the Abp1 protein, that both binds the mating-type region (Aguilar-Arnal et al. 2008) and regulates Swi2 transcription (Yu et al. 2012), Set1C and Brl2 might both act directly in the mating-type region and indirectly by controlling the production of Swi2 or other factors. In the mating-type region, some effects might be due to Set1C and Brl2 modulating Clr4-dependent methylation of histone H3K9.
A different mechanism for the directionality of mating-type switching in S. cerevisiae
The picture that emerges for the donor choice mechanism in S. pombe differs from donor preference in S. cerevisiae, as do other aspects of mating-type switching. The gene conversion of the MAT locus in S. cerevisiae are not dependent on passage of a replication fork through a nicked MAT locus as in S. pombe; instead, recombination is initiated by the HO endonuclease that creates a double-strand break at its recognition site at MATa or MATα [reviewed by Haber (2016), Lee and Haber (2015); Fig. 1]. Normally, HO endonuclease is expressed at the end of G1, after passage through “start,” but cells have entered S phase before recombination has been completed (Haber 2016). However, MAT switching can be induced both in cells arrested and maintained prior to S phase, when Cdc7 is inactivated (Hicks et al. 2011), or in G2/M-arrested cells (Wang et al. 2004), by expressing the HO endonuclease gene under control of a galactose-inducible promoter.
HO cleavage occurs close to the junction between the Ya- or Yα-specific sequences and the Z1 and Z2 homology regions shared between MAT and HML or HMR (Fig. 2). 5′ to 3′ resection of the DSB ends allows Rad51 protein to bind to the single-stranded DNA to promote strand invasion, copying of the donor Y region and termination in the longer shared homology regions W and X. Although HML shares more homology with MAT than does HMR, the 230 bp Z1 region is sufficient to yield nearly 100% recombination (Mehta et al. 2017).
As with S. pombe, a cell-type-specific donor selection mechanism can be inferred from the high efficiency of switching to the opposite mating-type in switching-competent cells, in this case, mother cells (Hicks and Herskowitz 1976; Strathern and Herskowitz 1979). The HML locus is preferentially selected in a-mating cells and the HMR locus is chosen in MATα cells. As in S. pombe, donor preference is not dependent on the specific mating-type sequences in the donor locus, as the same preference is seen when both donor loci contain Yα or when HML contains Ya sequences and HMR carries Yα (Klar et al. 1982; Rine et al. 1981). Both HML and HMR are maintained as short, heterochromatic, non-transcribed regions (Fig. 2), but unlike in S. pombe, the two loci are located far away from MAT and from each other, near the two telomeres of the same chromosome. MAT switching, therefore, requires long-range ectopic interactions between loci that are about 200 or 100 kb away from the HO-cut MAT locus (Fig. 1). As in S. pombe, the silenced donors have deacetylated histones; however, the histone H3K9 methylation pathway does not exist in S. cerevisiae and thus does not contribute to gene silencing or donor selection (Fig. 2). Instead, silencing of HML and HMR relies entirely on the Sir2 histone deacetylase pathway, which plays a less significant role for silencing in S. pombe.
Donor preference is controlled from a distance by a cell-type-specific Recombination Enhancer (RE), the ~ 750-bp multipartite RE element located 17 kb away from HML towards the centromere (Wu and Haber 1996) (Fig. 3). Sequences closer to the donor loci can be deleted with no effect on donor preference (Weiler and Broach 1992; Wu and Haber 1995). When RE is deleted, donor preference shifts from ~ 90% HML usage to less than 10%. In MATa cells, RE is bound by the Mcm1 protein (Szeto et al. 1997), the dimeric transcription factor Swi4/Swi6 (Coic et al. 2006) and multiple copies of the forkhead protein Fkh1 (Sun et al. 2002). Mechanistically, RE acts primarily through the several copies of Fkh1 bound to RE (Li et al. 2012). RE can be deleted and replaced by LexA binding sites to which LexA-Fkh1 (or simply the phospho-threonine binding domain of Fkh1) is bound. Fkh1 apparently binds to phospho-threonines produced by a protein kinase on proteins bound at or near the DSB, so that HML is effectively tethered ~ 20 kb from the DSB, via RE, while HMR remains nearly 100 kb distant (Avsaroglu et al. 2016). One phosphorylated protein, Mph1 (Dummer et al. 2016) has been implicated, but the effect of deleting Mph1 is much less than creating a Fkh1-R80A mutation that abolishes the phospho-threonine-binding of Fkh1 (Li et al. 2012). One protein kinase implicated in this process is casein kinase II (Ck2), but its targets have not been identified and its absence does not have as severe an effect as deleting Fkh1 (Li et al. 2012).
This proposed mechanism of action for RE, induced proximity of donor and recipient loci, differs from the facilitation of strand exchange proposed in S. pombe for SRE2 and SRE3, but might correspond to a function of SRE2 and SRE3 that still remains to be identified. However, a key difference between budding and fission yeast is that in the case of S. cerevisiae donor preference remains in full force even when one donor or both donors are unsilenced (Coic et al. 2011). Active RE is a “portable” enhancer of recombination, such that insertion of this short DNA sequence in different chromosomal locations will enhance the use of an ectopic donor even during interchromosomal recombination between unsilenced sequences that are unrelated to MAT (Lee et al. 2016; Roy et al. 2018). SRE2 or SRE3 are also to some extent ‘portable’ in that they remain active when their position is changed within the mating-type region (Jakociunas et al. 2013), but whether they function at other chromosomal locations to affect other DSB induced recombination events has not been tested.
In MATα cells, the MATα-encoded α2 protein dimerizes with Mcm1 and inactivates RE (Szeto et al. 1997); a change in chromatin structure is induced and the bound transcription factors are displaced (Weiss and Simpson 1997). RE is thereby inactivated in MATα cells and the default HMRa donor is chosen instead of HMLα. The preferential use of HMR over HML in MATα cells (or in MATa cells deleted for RE) is stronger than one would predict as a function of their relative distances to MAT (Avsaroglu et al. 2016), raising the possibility that there could be an unknown HMR-adjacent enhancer. An alternative possibility, based on the finding that adjacent to RE is a second Matα2-Mcm1 binding site (Szeto et al. 1997), is that RE could be tethered to another site, preventing it—and thus HML—from interacting with MATα. Indeed, artificially tethering a set of LacO repeats between HMLα and RE to the nuclear periphery by a LacI::FFAT domain does constrain the use of HML (Avsaroglu et al. 2014).
In MATa/MATα diploid cells, Matα2 and the MATa-encoded a1 protein form a heterodimer, which represses haploid-specific genes including the HO gene (Haber 2016). Therefore, switching is repressed in diploids. However, in S. pombe, diploids cells can switch mating-type on either chromosome (Egel 1984b; Egel and Eie 1987).
Cell fate in the absence of donors
Another striking difference between mating-type switching in budding and fission yeasts is the consequence of deleting both donor sequences when a DSB is induced. Induction of HO in an hml∆ hmr∆ strain results in near-total lethality in S. cerevisiae (Klar, et al. 1984), with about 0.2% of cells surviving by nonhomologous end-joining events that mutate the cleavage site (Moore and Haber 1996; Wu et al. 1996). In contrast, S. pombe cells deleted for both mat2-P and mat3-M but competent to undergo switching are viable (Klar and Miglio 1986), though their viability remains dependent on the presence of homologous recombination proteins such as Rad51 or SpRad22 (ScRad52) (Roseaulin et al. 2008). In the absence of the two donors, repair occurs by sister-chromatid recombination with the unbroken mat1 locus (since the DSB arises by replication through a single-strand nick). These results raise another question about the role of heterochromatin and the SRE loci in the repair of the mat1 DSB: why is sister-chromatid repair not always favored as the means of repair, since there is far more homology on both sides of the DSB than the remarkably short 59-bp H1 region that is used to initiate switching from the donors. These results suggest another role for the chromatin structure of the mating-type cassettes, to prevent sister-chromatid repair. It is interesting to note that, in budding yeast, an HO-induced DSB leads to the recruitment of cohesion around the break, which would promote sister-chromatid recombination (Ström et al. 2004; Ünal et al. 2004). How cohesion recruitment is affected by heterochromatin in the mating-type region in fission yeast would be worth examining. Interestingly, the switch-activating protein Sap1 that binds DNA elements at mat1 that are required for mating-type switching, is also present at IR-L and IR-R (Raimondi et al. 2018) and has been proposed to play a role in cohesion (de Lahondes et al. 2003).
Inhibitory versus stimulatory roles of heterochromatin on recombination
Some forms of recombination are inhibited by heterochromatin. The HML and HMR loci are protected from cleavage by the HO endonuclease (Strathern et al. 1982; White and Haber 1990), but they are cleaved in sir mutants and can then act as gene conversion recipients from an uncleavable MAT locus (Haber et al. 1980; Klar et al. 1981). In S. pombe, heterochromatin prevents both mitotic and meiotic recombination events. In meiosis, the occurrence of crossovers (CO) is repressed more than a thousand fold in the mat2-P - mat3-M interval (Egel 1984a, b); this block is released in heterochromatin mutants (Egel et al. 1989; Klar and Bonaduce 1991; Lorentz et al. 1992; Thon and Klar 1992, Thon and Klar 1994) or by deleting the heterochromatin nucleation center cenH located between mat2-P and mat3-M (Grewal and Klar 1997; Klar and Miglio 1986). In vegetative cells, chromosomal integration of transformed DNA is inefficient in the mating-type region and more efficient in heterochromatin mutants (Thon and Klar 1993). These recombination blocks stand in sharp contrast to the efficiency with which the silent donor loci are copied during mating-type switching. Recombination blocks also occur in pericentromeric heterochromatin. In meiosis, heterochromatin prevents COs in centromeric regions in S. pombe (Ellermeier et al. 2010, Fowler, et al. 2014), Arabidopsis (Underwood, et al. 2018) and other eukaryotes (reviewed by Nambiar and Smith 2016). In S. cerevisiae, meiotic recombination is also repressed near centromeres and in subtelomeric regions (Barton et al. 2008). There is a positive correlation between the location of meiotic hotspots and open chromatin (Berchowitz et al. 2009). The inhibitions have an important biological function, because COs too close to centromeres would interfere with chromosome segregation, or lead to gross genomic rearrangements if the COs occurred between common repeated sequences found on different chromosomes.
The mechanistic bases behind the inhibitions are only partly known. One level of control is through the inhibition of DSB formation. Heterochromatin prevents cutting by many restriction enzymes in addition to the HO endonuclease in the nuclei of mitotically dividing cells, suggesting the existence of steric hindrances (Loo and Rine 1994). Moreover, nucleases that gain close access to DNA might also be inactivated. For instance, in S. cerevisiae, tethering the meiotic nuclease Spo11 to various chromosomal locations sometimes results in DSBs but not always (Murakami and Nicolas 2009; Robine et al. 2007). Cohesins can have a determinant role (Nambiar and Smith 2018). In S. pombe meiosis, the mitotic cohesin subunit Psc3 is recruited by pericentromeric heterochromatin instead of the meiotic-specific cohesin Rec11 found in chromosome arms. The absence of Rec11 precludes activation of the Rec12(Spo11) pathway, of which Rec11 is an early effector. Thus, pericentromeric heterochromatin prevents DSB formation by Rec12(Spo11) and CO formation (Ellermeier et al. 2010; Fowler et al. 2014; Nambiar and Smith 2018).
One possible explanation for the differential effects of heterochromatin on different forms of recombination would be that nuclease access is prevented but that cells have the necessary chromatin remodelers to allow these same regions to be used as efficient donors (Chai et al. 2005; Hicks et al. 2011; Kent et al. 2007; van Attikum et al. 2004). Heterochromatin might also channel recombination to pathways that do not result in COs, this might be achieved on occasion through the use of distinct mediators of recombination that have the capacity of dictating recombination outcomes (Akamatsu et al. 2007). Schizosaccharomyces pombe mating-type switching shows that chromatin structure can affect the choice of recombination accessory factors; for example the poor switching of Swi2 and Swi5 mutants is significantly suppressed by the absence of Swi6 (Jia et al. 2004; Matsuda et al. 2011) indicating heterochromatin normally prohibits the use of alternate facilitators of strand exchange.
Perspectives
Genetic analyses of mating-type switching have identified a number of elements capable of conditionally imposing strong biases to recombination, some of which should be amenable to detailed in vivo monitoring after synchronized DSB formation as well as in vitro characterization. Might SRE2 and SRE3 facilitate strand exchange in vitro and what is their dependency on recombinases and accessory factors? In the future, genetic modifications of histones and biochemical assays using nucleosomal DNA might allow sorting out the effects of various histone modifications and protein complexes, to improve our understanding of how homologous recombination can be enhanced or suppressed.
References
Aboussekhra A, Chanet R, Adjiri A, Fabre F (1992) Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol Cell Biol 12:3224–3234. https://doi.org/10.1128/mcb.12.7.3224
Aguilar-Arnal L, Marsellach FX, Azorin F (2008) The fission yeast homologue of CENP-B, Abp1, regulates directionality of mating-type switching. EMBO J 27:1029–1038. https://doi.org/10.1038/emboj.2008.53
Akamatsu Y, Dziadkowiec D, Ikeguchi M, Shinagawa H, Iwasaki H (2003) Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc Natl Acad Sci USA 100:15770–15775. https://doi.org/10.1073/pnas.2632890100
Akamatsu Y, Tsutsui Y, Morishita T, Siddique MS, Kurokawa Y, Ikeguchi M, Yamao F, Arcangioli B, Iwasaki H (2007) Fission yeast Swi5/Sfr1 and Rhp55/Rhp57 differentially regulate Rhp51-dependent recombination outcomes. EMBO J 26:1352–1362. https://doi.org/10.1038/sj.emboj.7601582
Arcangioli B, de Lahondes R (2000) Fission yeast switches mating type by a replication-recombination coupled process. EMBO J 19:1389–1396. https://doi.org/10.1093/emboj/19.6.1389
Argunhan B, Murayama Y, Iwasaki H (2017) The differentiated and conserved roles of Swi5-Sfr1 in homologous recombination. Febs Lett 591:2035–2047. https://doi.org/10.1002/1873-3468.12656
Avsaroglu B, Bronk G, Gordon-Messer S, Ham J, Bressan DA, Haber JE, Kondev J (2014) Effect of chromosome tethering on nuclear organization in yeast. PLoS One 9:e102474. https://doi.org/10.1371/journal.pone.0102474
Avsaroglu B, Bronk G, Li K, Haber JE, Kondev J (2016) Chromosome-refolding model of mating-type switching in yeast. Proc Natl Acad Sci U S A. https://doi.org/10.1073/pnas.1607103113
Barton AB, Pekosz MR, Kurvathi RS, Kaback DB (2008) Meiotic recombination at the ends of chromosomes in Saccharomyces cerevisiae. Genetics 179:1221–1235. https://doi.org/10.1534/genetics.107.083493
Berchowitz LE, Hanlon SE, Lieb JD, Copenhaver GP (2009) A positive but complex association between meiotic double-strand break hotspots and open chromatin in Saccharomyces cerevisiae. Genome Res 19:2245–2257. https://doi.org/10.1101/gr.096297.109
Chai B, Huang J, Cairns BR, Laurent BC (2005) Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev 19:1656–1661. https://doi.org/10.1101/gad.1273105
Coic E, Sun K, Wu C, Haber JE (2006) Cell cycle-dependent regulation of Saccharomyces cerevisiae donor preference during mating-type switching by SBF (Swi4/Swi6) and Fkh1. Mol Cell Biol 26:5470–5480. https://doi.org/10.1128/MCB.02443-05
Coic E, Martin J, Ryu T, Tay SY, Kondev J, Haber JE (2011) Dynamics of homology searching during gene conversion in Saccharomyces cerevisiae revealed by donor competition. Genetics 189:1225–1233. https://doi.org/10.1534/genetics.111.132738
Dalgaard JZ, Klar AJ (2001) A DNA replication-arrest site RTS1 regulates imprinting by determining the direction of replication at mat1 in S. pombe. Genes Dev 15:2060–2068. https://doi.org/10.1101/gad.200801
de Lahondes R, Ribes V, Arcangioli B (2003) Fission yeast Sap1 protein is essential for chromosome stability. Eukaryot Cell 2:910–921
Dummer AM, Su Z, Cherney R, Choi K, Denu J, Zhao X, Fox CA (2016) Binding of the Fkh1 forkhead associated domain to a phosphopeptide within the Mph1 DNA helicase regulates mating-type switching in budding yeast. PLoS Genet 12:e1006094. https://doi.org/10.1371/journal.pgen.1006094
Egel R (1984a) Two tightly linked silent cassettes in the mating-type region of Schizosaccharomyces pombe. Curr Genet 8:199–203. https://doi.org/10.1007/BF00417816
Egel R (1984b) The pedigree pattern of mating-type switching in Schizosaccharomyces pombe. Curr Genet 8:205–210. https://doi.org/10.1007/BF00417817
Egel R, Eie B (1987) Cell Lineage Asymmetry in Schizosaccharomyces-Pombe – Unilateral Transmission of a High-Frequency State for Mating-Type Switching in Diploid Pedigrees. Curr Genet 12: 429–433 https://doi.org/10.1007/Bf00434820
Egel R, Beach DH, Klar AJ (1984) Genes required for initiation and resolution steps of mating-type switching in fission yeast. Proc Natl Acad Sci USA 81:3481–3485
Egel R, Willer M, Nielsen O (1989) Unblocking of meiotic crossing-over between the silent mating-type cassettes of fission yeast, conditioned by the recessive, pleiotropic mutant Rik1. Curr Genet 15:407–410. https://doi.org/10.1007/Bf00376796
Ellermeier C, Higuchi EC, Phadnis N, Holm L, Geelhood JL, Thon G, Smith GR (2010) RNAi and heterochromatin repress centromeric meiotic recombination. Proc Natl Acad Sci USA 107:8701–8705. https://doi.org/10.1073/pnas.0914160107
Fowler KR, Sasaki M, Milman N, Keeney S, Smith GR (2014) Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome. Genome Res 24:1650–1664. https://doi.org/10.1101/gr.172122.114
Grewal SI, Klar AJ (1997) A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics 146:1221–1238
Haber (Haber (2016) JE (2016) A life investigating pathways that repair broken chromosomes. Annu Rev Genet 50: 1–28 https://doi.org/10.1146/annurev-genet-120215-035043
Haber JE, Mascioli DW, Rogers DT (1980) Illegal transposition of mating-type genes in yeast. Cell 20:519–528
Hansen KR, Hazan I, Shanker S, Watt S, Verhein-Hansen J, Bahler J, Martienssen RA, Partridge JF, Cohen A, Thon G (2011) H3K9me-independent gene silencing in fission yeast heterochromatin by Clr5 and histone deacetylases. PLoS Genet 7:e1001268. https://doi.org/10.1371/journal.pgen.1001268
Haruta N, Kurokawa Y, Murayama Y, Akamatsu Y, Unzai S, Tsutsui Y, Iwasaki H (2006) The Swi5-Sfr1 complex stimulates Rhp51/Rad51- and Dmc1-mediated DNA strand exchange in vitro. Nat Struct Mol Biol 13:823–830. https://doi.org/10.1038/nsmb1136
Hicks JB, Herskowitz I (1976) Interconversion of yeast mating types I. Direct observations of the action of the homothallism (HO) Gene. Genetics 83:245–258
Hicks WM, Yamaguchi M, Haber JE (2011) Real-time analysis of double-strand DNA break repair by homologous recombination. Proc Natl Acad Sci USA 108:3108–3115. https://doi.org/10.1073/pnas.1019660108
Ito K, Murayama Y, Takahashi M, Iwasaki H (2018) Two three-strand intermediates are processed during Rad51-driven DNA strand exchange. Nat Struct Mol Biol 25:29–36. https://doi.org/10.1038/s41594-017-0002-8
Jakociunas T, Holm LR, Verhein-Hansen J, Trusina A, Thon G (2013) Two portable recombination enhancers direct donor choice in fission yeast heterochromatin. PLoS Genet 9:e1003762. https://doi.org/10.1371/journal.pgen.1003762
Jia S, Yamada T, Grewal SI (2004) Heterochromatin regulates cell type-specific long-range chromatin interactions essential for directed recombination. Cell 119:469–480. https://doi.org/10.1016/j.cell.2004.10.020
Kent NA, Chambers AL, Downs JA (2007) Dual chromatin remodeling roles for RSC during DNA double strand break induction and repair at the yeast MAT locus. J Biol Chem 282:27693–27701. https://doi.org/10.1074/jbc.M704707200
Klar AJS (1990) The developmental fate of fission yeast-cells is determined by the pattern of inheritance of parental and grandparental DNA Strands. Embo Journal 9:1407–1415. https://doi.org/10.1002/j.1460-2075.1990.tb08256.x
Klar AJ, Bonaduce MJ (1991) swi6, a gene required for mating-type switching, prohibits meiotic recombination in the mat2-mat3 “cold spot” of fission yeast. Genetics 129:1033–1042
Klar AJ, Miglio LM (1986) Initiation of meiotic recombination by double-strand DNA breaks in S. pombe. Cell 46:725–731. https://doi.org/10.1016/0092-8674(86)90348-x
Klar AJ, Strathern JN, Hicks JB (1981) A position-effect control for gene transposition: state of expression of yeast mating-type genes affects their ability to switch. Cell 25:517–524. https://doi.org/10.1016/0092-8674(81)90070-2
Klar AJS, Hicks JB, Strathern JN (1982) Directionality of yeast mating-type interconversion. Cell 28:551–561. https://doi.org/10.1016/0092-8674(82)90210-0
Klar AJ, Strathern JN, Abraham JA (1984) Involvement of double-strand chromosomal breaks for mating-type switching in Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol 49:77–88. https://doi.org/10.1101/sqb.1984.049.01.011
Klar AJ, Ishikawa K, Moore S (2014) A unique DNA recombination mechanism of the mating/cell-type switching of fission yeasts: a review. Microbiol Spectr 210.1128/microbiolspec.MDNA3-0003-2014
Kostriken R, Strathern JN, Klar AJ, Hicks JB, Heffron F (1983) A site-specific endonuclease essential for mating-type switching in Saccharomyces cerevisiae. Cell 35:167–174. https://doi.org/10.1016/0092-8674(83)90219-2
Kurokawa Y, Murayama Y, Haruta-Takahashi N, Urabe I, Iwasaki H (2008) Reconstitution of DNA strand exchange mediated by Rhp51 recombinase and two mediators. PLoS Biol 6:e88. https://doi.org/10.1371/journal.pbio.0060088
Lee CS, Haber JE (2015) Mating-type gene switching in Saccharomyces cerevisiae. Microbiol Spectr 3:MDNA3-0013-2014. https://doi.org/10.1128/microbiolspec.MDNA3-0013-2014
Lee CS, Wang RW, Chang HH, Capurso D, Segal MR, Haber JE (2016) Chromosome position determines the success of double-strand break repair. Proc Natl Acad Sci USA 113:E146–E154. https://doi.org/10.1073/pnas.1523660113
Li J, Coic E, Lee K, Lee CS, Kim JA, Wu Q, Haber JE (2012) Regulation of budding yeast mating-type switching donor preference by the FHA domain of Fkh1. PLoS Genet 8:e1002630. https://doi.org/10.1371/journal.pgen.1002630
Loo S, Rine J (1994) Silencers and domains of generalized repression. Science 264:1768–1771. https://doi.org/10.1126/science.8209257
Lorentz A, Heim L, Schmidt H (1992) The switching gene Swi6 affects recombination and gene-expression in the mating-type region of Schizosaccharomyces pombe. Mol Gen Genet 233:436–442. https://doi.org/10.1007/bf00265441
Maki T, Ogura N, Haber JE, Iwasaki H, Thon G (2018) New insights into donor directionality of mating-type switching in Schizosaccharomyces pombe. PLoS Genet 14:e1007424. https://doi.org/10.1371/journal.pgen.1007424
Malone RE, Esposito RE (1980) The RAD52 gene is required for homothallic interconversion of mating types and spontaneous mitotic recombination in yeast. Proc Natl Acad Sci USA 77:503–507. https://doi.org/10.1073/pnas.77.1.503
Materne P, Anandhakumar J, Migeot V, Soriano I, Yague-Sanz C, Hidalgo E, Mignion C, Quintales L, Antequera F, Hermand D (2015) Promoter nucleosome dynamics regulated by signalling through the CTD code. Elife 4:e09008. https://doi.org/10.7554/eLife.09008
Materne P, Vazquez E, Sanchez M, Yague-Sanz C, Anandhakumar J, Migeot V, Antequera F, Hermand D (2016) Histone H2B ubiquitylation represses gametogenesis by opposing RSC-dependent chromatin remodeling at the ste11 master regulator locus. Elife. https://doi.org/10.7554/eLife.13500
Matsuda E, Sugioka-Sugiyama R, Mizuguchi T, Mehta S, Cui B, Grewal SI (2011) A homolog of male sex-determining factor SRY cooperates with a transposon-derived CENP-B protein to control sex-specific directed recombination. Proc Natl Acad Sci U S A 108:18754–18759. https://doi.org/10.1073/pnas.1109988108
Mehta A, Beach A, Haber JE (2017) Homology requirements and competition between gene conversion and break-induced replication during double-strand break repair. Mol Cell 65:515–526 e513. https://doi.org/10.1016/j.molcel.2016.12.003
Mikheyeva IV, Grady PJ, Tamburini FB, Lorenz DR, Cam HP (2014) Multifaceted genome control by Set1 dependent and independent of H3K4 methylation and the Set1C/COMPASS complex. PLoS Genet 10:e1004740. https://doi.org/10.1371/journal.pgen.1004740
Miyata H, Miyata M (1981) Mode of Conjugation in Homothallic Cells of Schizosaccharomyces-Pombe. J Gen Appl Microbiol 27:365–371. https://doi.org/10.2323/jgam.27.365
Moore JK, Haber JE (1996) Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol 16:2164–2173
Murakami H, Nicolas A (2009) Locally, meiotic double-strand breaks targeted by Gal4BD-Spo11 occur at discrete sites with a sequence preference. Mol Cell Biol 29:3500–3516. https://doi.org/10.1128/Mcb.00088-09
Nambiar M, Smith GR (2016) Repression of harmful meiotic recombination in centromeric regions. Semin Cell Dev Biol 54:188–197. https://doi.org/10.1016/j.semcdb.2016.01.042
Nambiar M, Smith GR (2018) Pericentromere-specific cohesin complex prevents meiotic pericentric DNA double-strand breaks and lethal crossovers. Mol Cell 71:540. https://doi.org/10.1016/j.molcel.2018.06.035
Nasmyth K (1983) Molecular analysis of a cell lineage. Nature 302:670–676. https://doi.org/10.1038/302670a0
Noma K, Allis CD, Grewal SIS (2001) Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293:1150–1155. https://doi.org/10.1126/science.1064150
Ostermann K, Lorentz A, Schmidt H (1993) The Fission Yeast Rad22 Gene, Having a Function in Mating-Type Switching and Repair of DNA Damages, Encodes a Protein Homolog to Rad52 of Saccharomyces-Cerevisiae. Nucleic Acids Res 21:5940–5944. https://doi.org/10.1093/nar/21.25.5940
Raimondi C, Jagla B, Proux C, Waxin H, Gangloff S, Arcangioli B (2018) Molecular signature of the imprintosome complex at the mating-type locus in fission yeast. Microb Cell 5:169–183. https://doi.org/10.15698/mic2018.04.623
Rine J, Jensen R, Hagen D, Blair L, Herskowitz I (1981) Pattern of switching and fate of the replaced cassette in yeast mating-type interconversion. Cold Spring Harb Symp Quant Biol 45 Pt 2:951–960. https://doi.org/10.1101/sqb.1981.045.01.112
Robine N, Uematsu N, Amiot F, Gidrol X, Barillot E, Nicolas A, Borde V (2007) Genome-wide redistribution of meiotic double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol 27:1868–1880. https://doi.org/10.1128/Mcb.02063-06
Roseaulin L, Yamada Y, Tsutsui Y, Russell P, Iwasaki H, Arcangioli B (2008) Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J 27:1378–1387. https://doi.org/10.1038/emboj.2008.65
Roy KR, Smith JD, Vonesch SC, Lin G, Tu CS, Lederer AR, Chu A, Suresh S, Nguyen M, Horecka J, Tripathi A, Burnett WT, Morgan MA, Schulz J, Orsley KM, Wei W, Aiyar RS, Davis RW, Bankaitis VA, Haber JE, Salit ML, St Onge RP, Steinmetz LM (2018) Multiplexed precision genome editing with trackable genomic barcodes in yeast. Nat Biotechnol 36:512–520. https://doi.org/10.1038/nbt.4137
Strathern JN, Herskowitz I (1979) Asymmetry and directionality in production of new cell types during clonal growth: the switching pattern of homothallic yeast. Cell 17:371–381. https://doi.org/10.1016/0092-8674(79)90163-6
Strathern JN, Klar AJS, Hicks JB, Abraham JA, Ivy JM, Nasmyth KA, Mcgill C (1982) Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the mat locus. Cell 31:183–192. https://doi.org/10.1016/0092-8674(82)90418-4
Ström L, Lindroos HB, Shirahige K, Sjögren C (2004) Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell 16:1003–1015. https://doi.org/10.1016/j.molcel.2004.11.026
Sun K, Coic E, Zhou Z, Durrens P, Haber JE (2002) Saccharomyces forkhead protein Fkh1 regulates donor preference during mating-type switching through the recombination enhancer. Genes Dev 16:2085–2096. https://doi.org/10.1101/gad.994902
Szeto L, Fafalios MK, Zhong H, Vershon AK, Broach JR (1997) alpha 2p controls donor preference during mating type interconversion in yeast by inactivating a recombinational enhancer of chromosome III. Genes Dev 11:1899–1911. https://doi.org/10.1101/gad.11.15.1899
Thon G, Klar AJ (1992) The clr1 locus regulates the expression of the cryptic mating-type loci of fission yeast. Genetics 131:287–296
Thon G, Klar AJ (1993) Directionality of fission yeast mating-type interconversion is controlled by the location of the donor loci. Genetics 134:1045–1054
Thon GC, Klar AAJ (1994) Three additional linkage groups that repress transcription and meiotic recombination in the mating-type region of Schizosaccharomyces pombe. Genetics 138:29–38
Thon G, Verhein-Hansen J (2000) Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics 155:551–568
Thon G, Hansen KR, Altes SP, Sidhu D, Singh G, Verhein-Hansen J, Bonaduce MJ, Klar AJ (2005) The Clr7 and Clr8 directionality factors and the Pcu4 cullin mediate heterochromatin formation in the fission yeast Schizosaccharomyces pombe. Genetics 171:1583–1595. https://doi.org/10.1534/genetics.105.048298
Ünal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D (2004) DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell 16:991–1002. https://doi.org/10.1016/j.molcel.2004.11.027
Underwood CJ, Choi K, Lambing C, Zhao X, Serra H, Borges F, Simorowski J, Ernst E, Jacob Y, Henderson IR, Martienssen RA (2018) Epigenetic activation of meiotic recombination near Arabidopsis thaliana centromeres via loss of H3K9me2 and non-CG DNA methylation. Genome Res 28:519–531. https://doi.org/10.1101/gr.227116.117
van Attikum H, Fritsch O, Hohn B, Gasser SM (2004) Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119:777–788. https://doi.org/10.1016/j.cell.2004.11.033
Wang X, Ira G, Tercero JA, Holmes AM, Diffley JF, Haber JE (2004) Role of DNA replication proteins in double-strand break-induced recombination in Saccharomyces cerevisiae. Mol Cell Biol 24:6891–6899. https://doi.org/10.1128/MCB.24.16.6891-6899.2004
Weiler KS, Broach JR (1992) Donor locus selection during Saccharomyces cerevisiae mating type interconversion responds to distant regulatory signals. Genetics 132:929–942
Weiss K, Simpson RT (1997) Cell type-specific chromatin organization of the region that governs directionality of yeast mating type switching. EMBO J 16:4352–4360. https://doi.org/10.1093/emboj/16.14.4352
White CI, Haber JE (1990) Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J 9:663–673. https://doi.org/10.1002/j.1460-2075.1990.tb08158.x
Wu X, Haber JE (1995) MATa donor preference in yeast mating-type switching: activation of a large chromosomal region for recombination. Genes Dev 9:1922–1932. https://doi.org/10.1101/gad.9.15.1922
Wu X, Haber JE (1996) A 700 bp cis-acting region controls mating-type dependent recombination along the entire left arm of yeast chromosome III. Cell 87:277–285. https://doi.org/10.1016/s0092-8674(00)81345-8
Wu X, Moore JK, Haber JE (1996) Mechanism of MAT alpha donor preference during mating-type switching of Saccharomyces cerevisiae. Mol Cell Biol 16:657–668. https://doi.org/10.1128/mcb.16.2.657
Yu C, Bonaduce MJ, Klar AJ (2012) Going in the right direction: mating-type switching of Schizosaccharomyces pombe is controlled by judicious expression of two different swi2 transcripts. Genetics 190:977–987. https://doi.org/10.1534/genetics.111.137109
Zofall M, Grewal SIS (2007) HULC, a histone H2B ubiquitinating complex, modulates heterochromatin independent of histone methylation in fission yeast. J Biol Chem 282:14065–14072. https://doi.org/10.1074/jbc.M700292200
Acknowledgements
This work was funded by the Japan Society Promotion of Science Grants-in-Aid or Scientific Research (A) (18H03985) and on Innovative Areas (15H05974) to Hiroshi Iwasaki, NIH grant R35 127029 to James Haber and by the Lundbeck Foundation grant R9-2007-867 and the Carlsberg Foundation grants 2012_01_0598 and CF15-0853 to Genevieve Thon.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Kupiec.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Thon, G., Maki, T., Haber, J.E. et al. Mating-type switching by homology-directed recombinational repair: a matter of choice. Curr Genet 65, 351–362 (2019). https://doi.org/10.1007/s00294-018-0900-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-018-0900-2