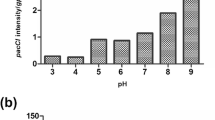
Abstract
A cluster of pathogenicity genes (PEP1, PEP2, PDA1, PEP5), termed the pea pathogenicity (PEP) cluster and located on a 1.6-Mb conditionally dispensable (CD) chromosome, was identified in the fungal pathogen Nectria haematococca. Studies determined that the expression of PDA1 is induced in both infected pea tissues and in vitro by the phytoalexin pisatin. The present study reports the use of real-time quantitative RT-PCR to monitor the expression of each PEP gene and PDA1. In mycelia actively growing in culture, the mRNA levels of PEP1, PEP5 and PDA1 were very low and the PEP2 transcript was undetectable. In planta, PDA1 and PEP2 were strongly induced, while PEP1 and PEP5 were moderately induced. Starvation slightly enhanced the expression of PEP1, PDA1 and PEP5, while the expression of PEP2 remained undetectable. Exposure to pisatin in culture stimulated the expression of PDA1 and each PEP gene to a similar level as occurred in planta. In addition, all four pathogenicity genes displayed similar temporal patterns of expression in planta and in vitro, consistent with a coordinated regulation of these genes by pisatin during pea pathogenesis. In the flanking regions of the PEP cluster, six open reading frames (ORFs) were identified and all were expressed during infection of pea. Comparison of the codon preferences of these ORFs and seven additional genes from CD chromosomes with the codon preferences of 21 genes from other chromosomes revealed there is a codon bias that correlates with the source of the genes. This difference in codon bias is consistent with the hypothesis that genes on the CD chromosome have a different origin from genes of normal chromosomes, suggesting that horizontal gene transfer may have played a role in the evolution of pathogenesis in N. haematococca.





Similar content being viewed by others
References
Ahn JH, Cheng YQ, Walton JD (2002) An extended physical map of the TOX2 locus of Cochliobolus carbonum required for biosynthesis of HC-toxin. Fungal Genet Biol 35:31–38
Baranowski JD; Davidson PM, Nagel CW, Branen AL (1980) Inhibition of Saccharomyces cerevisiae by naturally occurring hydroxycinnamates. J Food Sci 45:592–594
Black SD, Coon MJ (1987) Cytochromes P450: structure and function. Adv Enzymol 60:35–87
Clausen M, Lamb CJ, Megnet R, Doerner PW (1994) PAD1 encodes phenylacrylic acid decarboxylase which confers resistance to cinnamic acid in Saccharomyces cerevisiae. Gene 142:107–112
Covert SF (1998) Supernumerary chromosomes in filamentous fungi. Curr Genet 33:311–319
Covert SF, Enkerli J, Miao V PW, VanEtten HD (1996) A gene for maackiain detoxification from a dispensable chromosome of Nectria haematococca. Mol Gen Genet 251:397–406
Eddine AN, Hannemann F, Schafer W (2001) Cloning and expression analysis of NhL1, a gene encoding an extracellular lipase from the fungal pea pathogen Nectria haematococca MP VI (Fusarium solani f. sp. pisi) that is expressed in planta. Mol Genet Genomics 265:215–224
Enkerli J, Bhatt G, Covert SF (1997) Nht1, a transposable element cloned from a dispensable chromosome in Nectria haematococca. Mol Plant-Microbe Interact 10:742-749
Funnell DL, VanEtten HD (2002) Pisatin demethylase genes are on dispensable chromosomes while genes for pathogenicity on carrot and ripe tomato are on other chromosomes in Nectria haematococca. Mol Plant-Microbe Interact 15:840–846
Hacker J, Carniel E (2001) Ecological fitness, genomic islands and bacterial pathogenicity—a Darwinian view of the evolution of microbes. EMBO Rep 2:376–381
Hacker J, Kaper JB (2000) Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol 54:641–679
Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H (1997) Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol 23:1089-1097
Han Y, Liu X, Benny U, Kistler HC, VanEtten HD (2001) Gene determining pathogenicity to pea are clustered on a supernumerary chromosome in the fungal plant pathogen Nectria haematococca. Plant J 25:305–314
Hatta R, Ito K, Hosaki Y, Tanaka T, Tanaka A, Yamamoto M, Akimitsu K (2002) A conditionally dispensable chromosome controls host-specific pathogenicity in the fungal plant pathogen Alternaria alternata. Genetics 161:59–70
He CZ, Rusu AG, Poplawski AM, Irwin JAG, Manners JM (1998) Transfer of a supernumerary chromosome between vegetatively incompatible biotypes of the fungus Colletotrichum gloeosporioides. Genetics 150:1459-1466
Hirschi K, VanEtten HD (1996) Expression of the pisatin detoxifying gene (PDA) of Nectria haematococca in vitro and in planta. Mol Plant-Microbe Interact 9:483–491
Hueck CJ (1998) Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev 62:379–433
Huyan TV, Dahlbeck D, Staskawicz BJ (1989) Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245:1374–1377
Johnson LJ, Johnson RD, Akamatsu H, Salamiah A, Otani H, Kohmoto K, Kodama M (2001) Spontaneous loss of a conditionally dispensable chromosome from the Alternaria alternata apple pathotype leads to loss of toxin production and pathogenicity. Curr Genet 40:65–72
Kahmann R, Basse C (2001). Fungal gene expression during pathogenesis-related development and host plant colonization. Curr Opin Microbiol 4:374–380
Kaufman L, Rousseeuw PJ (1990) Finding groups in data: an introduction to cluster analysis. Wiley, New York
Khan K, Straney DC (1999) Regulatory signals influencing expression of the PDA1 gene of Nectria haematococca MPVI in culture and during pathogenesis of pea. Mol Plant-Microbe Interact 12:733-742
Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389-3402
Maloney AP, VanEtten HD (1994) A gene from the fungal plant pathogen Nectria haematococca that encodes the phytoalexin-detoxifying enzyme pisatin demethylase defines a new cytochrome P450 family. Mol Gen Genet 243:506-514
Miao VPW, Matthews DE, VanEtten HD (1991) Identification and chromosomal locations of a family of cytochrome P-450 genes for pisatin detoxification in the fungus Nectria haematococca. Mol Gen Genet 226:214-223
Miller JF, Mekalanos JJ, Falkow S (1989) Coordinate regulation and sensory transduction in the control of bacterial virulence. Science 243:916–921
Ochman H, Moran N (2001) Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292:1096–1098
Ochman H, Lawrence JG, Groisman EA (2000) Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304
Pieterse CMJ, Derksen AMCE, Folders J, Govers F (1994) Expression of the Phytophthora infestans ipiB and ipiO genes in planta and in vitro. Mol Gen Genet 244:269–277
Ragan MA (2001) Detection of lateral gene transfer among microbial genomes. Curr Opin Genet Dev 11:620–626.
Reimmann C, VanEtten HD (1994) Cloning and characterization of the PDA6-1 gene encoding a fungal cytochrome P-450 which detoxifies the phytoalexin pisatin from garden pea. Gene 146:221–226
Rose MS, Yun SH, Asvarak T, Lu SW, Yoder OC, Turgeon BG (2002) A decarboxylase encoded at the Cochliobolus heterostrophus translocation-associated Tox1B locus is required for polyketide (T-toxin) biosynthesis and high virulence on T-cytoplasm maize. Mol Plant-Microbe Interact 15:883–893
Rosewich UL, Kistler HC (2000) Role of horizontal gene transfer in the evolution of fungi. Annu Rev Phytopathol 38:325–363
Ruan Y, Straney DC (1996) Identification of elements in the PDA1 promoter of Nectria haematococca necessary for a high level transcription in vitro. Mol Gen Genet 250:29–38
Schmidt H, Zhang WL, Hemmrich U, Jelacic S, Brunder W, Tarr PI, Dobrindt U, Hacker J, Karch H (2001) Identification and characterization of a novel genomic island integrated at selC in locus of enterocyte effacement-negative, shiga toxin-producing Escherichia coli. Infect Immun 69:6863–6873
Stephans RB (1974) Mycological guidebook. University of Washington Press, Seattle, Wash.
Straney DC, VanEtten HD (1994) Characterization of the PDA1 promoter of Nectria haematococca and the identification of a region that binds a pisatin-responsive DNA binding factor. Mol Plant-Microbe Interact 7:256–266
Struyf A, Hubert M, Rousseeuw PJ (1997) Integrating robust clustering techniques in S-PLUS. Comput Stat Data Anal 26:17–37
Sullivan JT, Ronson CW (1998) Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc Natl Acad Sci USA 95:5145–5149
Teasdale J, Daniels D, Davis WC, Eddy R, Hadwiger LA (1974) Physiological and cytological similarities between disease resistance and cellular incompatibility responses. Plant Physiol 54:690–695
Tegtmeier KJ, VanEtten HD (1982) The role of pisatin tolerance and degradation in the virulence of Nectria haematococca on peas: a genetic analysis. Phytopathology 72:608–612
Temporini ED, VanEtten HD (2002) Distribution of the pea pathogenicity (PEP) genes in the fungus Nectria haematococca mating population VI. Curr Genet 41:107–114
Van den Ackerveken GFJM, Dunn RM, Cozjnsen TJ, Vossen P, Van den Broek HWJ, De Wit PJGM (1994) Nitrogen limitation induced expression of the avirulence gene avr9 in the tomato pathogen Cladosporium fulvum. Mol Gen Genet 243:277–295
VanEtten HD (1978) Identification of additional habitats of Nectria haematococca mating population VI. Phytopathology 68:1552–1556
VanEtten HD, Jorgensen S, Enkerli J, Covert SF (1998) Inducing the loss of conditionally dispensable chromosomes in Nectria haematococca during vegetative growth. Curr Genet 33:299–303
Walton JD (2000) Horizontal gene transfer and the evolution of secondary metabolite gene clusters in fungi: an hypothesis. Fungal Genet Biol 30:167–171
Wasmann CC, VanEtten HD (1996) Transformation-mediated chromosome loss and disruption of a gene for pisatin demethylase decrease the virulence of Nectria haematococca on pea. Mol Plant-Microbe Interact 9:793-803
Acknowledgements
This work was supported in part by grant 9905422 from the National Science Foundation, Division of Molecular and Cellular Biosciences. We appreciate the research advice and editorial help of Catherine Wasmann and Esteban Temporini and the technical assistance of Catherine Wasmann and Gerard White. We gratefully acknowledge technical advice from George Vandemark on the use of TaqMan technology for real-time quantitative PCR and thank David Galbraith for use of his Smart Cycler System.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Heitman
Rights and permissions
About this article
Cite this article
Liu, X., Inlow, M. & VanEtten, H.D. Expression profiles of pea pathogenicity (PEP) genes in vivo and in vitro, characterization of the flanking regions of the PEP cluster and evidence that the PEP cluster region resulted from horizontal gene transfer in the fungal pathogen Nectria haematococca . Curr Genet 44, 95–103 (2003). https://doi.org/10.1007/s00294-003-0428-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-003-0428-x