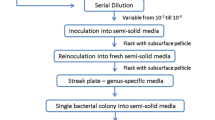
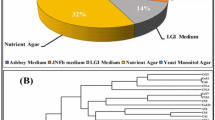
Abstract
Three microbial strains were isolated from the rhizosphere of alfalfa (Medicago sativa), grass mixture (Festuca rubra, 75 %; Lolium perenne, 20 %; Poa pratensis, 10 %), and rape (Brassica napus) on the basis of their high capacity to use crude oil as the sole carbon and energy source. These isolates used an unusually wide spectrum of hydrocarbons as substrates (more than 80), including n-alkanes with chain lengths ranging from C12 to C32, monomethyl- and monoethyl-substituted alkanes (C12–C23), n-alkylcyclo alkanes with alkyl chain lengths from 4 to 18 carbon atoms, as well as substituted monoaromatic and diaromatic hydrocarbons. These three strains were identified as Gordonia rubripertincta and Rhodococcus sp. SBUG 1968. During their transformation of this wide range of hydrocarbon substrates, a very large number of aliphatic, alicyclic, and aromatic acids was detected, 44 of them were identified by GC/MS analyses, and 4 of them are described as metabolites for the first time. Inoculation of plant seeds with these highly potent bacteria had a beneficial effect on shoot and root development of plants which were grown on oil-contaminated sand.


Similar content being viewed by others
References
Andreoni V, Bernasconi S, Colombo M, van Beilen JB, Cavalca L (2000) Detection of genes for alkane and naphthalene catabolism in Rhodococcus sp. strain 1BN. Environ Microbiol 2(5):572–577. doi:10.1046/j.1462-2920.2000.00134.x
Arenskötter M, Bröker D, Steinbüchel A (2004) Biology of the metabolically diverse genus Gordonia. Appl Environ Microbiol 70(6):3195–3204. doi:10.1128/aem. 70.6.3195-3204.2004
Atlas RM (1991) Microbial hydrocarbon degradation - bioremediation of oil spills. J Chem Technol Biotechnol 52(2):149–156
Atlas RM, Atlas MC (1991) Biodegradation of oil and bioremediation of oil spills. Curr Opin Biotechnol 2(3):440–443. doi:10.1016/s0958-1669(05)80153-3
Awe S, Mikolasch A, Hammer E, Schauer F (2008) Degradation of phenylalkanes and characterization of aromatic intermediates acting as growth inhibiting substances in hydrocarbon utilizing yeast Candida maltosa. Int Biodeterior Biodegrad 62(4):408–414. doi:10.1016/j.ibiod.2008.03.007
Awe S, Mikolasch A, Schauer F (2009) Formation of coumarines during the degradation of alkyl substituted aromatic oil components by the yeast Trichosporon asahii. Appl Microbiol Biotechnol 84(5):965–976. doi:10.1007/s00253-009-2044-2
Beam HW, Perry JJ (1974) Microbial degradation and assimilation of n-alkyl-substituted cycloparaffins. J Bacteriol 118(2):394–399
Bhatia M, Singh HD (1996) Biodegradation of commercial linear alkyl benzenes by Nocardia amarae. J Biosci 21(4):487–496. doi:10.1007/bf02703213
Blakley ER, Papish B (1982) The metabolism of cyclohexanecarboxylic acid and 3-cyclohexenecarboxylic acid by Pseudomonas putida. Can J Microbiol 28(12):1324–1329
Cirou A, Diallo S, Kurt C, Latour X, Faure D (2007) Growth promotion of quorum-quenching bacteria in the rhizosphere of Solanum tuberosum. Environ Microbiol 9(6):1511–1522. doi:10.1111/j.1462-2920.2007.01270.x
Clayton RA, Sutton G, Hinkle PS, Bult C, Fields C (1995) Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int J Syst Bacteriol 45(3):595–599
De Boer TD, Backer HJ (1956) Diazomethane. In: J LN (ed) Organic synthesis, vol 36. Wiley, New York, NY, pp 14–16
de Carvalho CCCR, da Fonseca MMR (2005a) Degradation of hydrocarbons and alcohols at different temperatures and salinities by Rhodococcus erythropolis DCL14. FEMS Microbiol Ecol 51(3):389–399. doi:10.1016/j.femsec.2004.09.010
de Carvalho CCCR, da Fonseca MMR (2005b) The remarkable Rhodococcus erythropolis. Appl Microbiol Biotechnol 67(6):715–726. doi:10.1007/s00253-005-1932-3
de Carvalho CCCR, Parreno-Marchante B, Neumann G, da Fonseca MMR, Heipieper HJ (2005) Adaptation of Rhodococcus erythropolis DCL14 to growth on n-alkanes, alcohols and terpenes. Appl Microbiol Biotechnol 67(3):383–388. doi:10.1007/s00253-004-1750-z
de Carvalho CCCR, Wick LY, Heipieper HJ (2009) Cell wall adaptations of planktonic and biofilm Rhodococcus erythropolis cells to growth on C5 to C16 n-alkane hydrocarbons. Appl Microbiol Biotechnol 82(2):311–320. doi:10.1007/s00253-008-1809-3
Diallo S, Crepin A, Barbey C, Orange N, Burini J-F, Latour X (2011) Mechanisms and recent advances in biological control mediated through the potato rhizosphere. FEMS Microbiol Ecol 75(3):351–364. doi:10.1111/j.1574-6941.2010.01023.x
Dua M, Singh A, Sethunathan N, Johri AK (2002) Biotechnology and bioremediation: successes and limitations. Appl Microbiol Biotechnol 59(2–3):143–152. doi:10.1007/s00253-002-1024-6
Dutta TK, Harayama S (2001) Biodegradation of n-alkylcycloalkanes and n-alkylbenzenes via new pathways in Alcanivorax sp. strain MBIC 4326. Appl Environ Microbiol 67(4):1970–1974. doi:10.1128/aem. 67.4.1970-1974.2001
Fedorak PM, Westlake DWS (1986) Fungal metabolism of n-alkylbenzenes. Appl Environ Microbiol 51(2):435–437
Fedorak PM, Payzant JD, Montgomery DS, Westlake DWS (1988) Microbial degradation of n-alkyl tetrahydrothiophenes found in petroleum. Appl Environ Microbiol 54(5):1243–1248
Fedorak PM, Coy DL, Peakman TM (1996) Microbial metabolism of some 2,5-substituted thiophenes. Biodegradation 7(4):313–327. doi:10.1007/bf00115745
Feinberg EL, Ramage PIN, Trudgill PW (1980) The degradation of n-alkylcycloalkanes by a mixed bacterial culture. J Gen Microbiol 121(DEC):507–511
Gailiūtė I, Kavaliauskė M, Aikaitė-Stanaitienė J (2011) Changes in total oil hydrocarbon composition during degradation with sorbent bacterial preparation. Biologija 57:70–77
Gallego S, Vila J, Tauler M, Nieto JM, Breugelmans P, Springael D, Grifoll M (2014) Community structure and PAH ring-hydroxylating dioxygenase genes of a marine pyrene-degrading microbial consortium. Biodegradation 25(4):543–556. doi:10.1007/s10532-013-9680-z
Gerhardt KE, Huang X-D, Glick BR, Greenberg BM (2009) Phytoremediation and rhizoremediation of organic soil contaminants: potential and challenges. Plant Sci 176(1):20–30. doi:10.1016/j.plantsci.2008.09.014
Graj W, Lisiecki P, Szulc A, Chrzanowski L, Wojtera-Kwiczor J (2013) Bioaugmentation with petroleum-degrading consortia has a selective growth-promoting impact on crop plants germinated in diesel oil-contaminated soil. Water Air Soil Pollut 224:1676. doi:10.1007/s11270-013-1676-0
Heipieper HJ (2007) Bioremediation of soils contaminated with aromatic compounds: Effects of rhizosphere, bioavailability, gene regulation and stress adaptation. In: Heipieper HJ (ed) Bioremediation of soils contaminated with aromatic compounds. NATO Science Series IV-Earth and Environmental Sciences, vol 76, pp 1–4
Herter S, Mikolasch A, Schauer F (2012) Identification of phenylalkane derivatives when Mycobacterium neoaurum and Rhodococcus erythropolis were cultured in the presence of various phenylalkanes. Appl Microbiol Biotechnol 93(1):343–355. doi:10.1007/s00253-011-3415-z
Hong SH, Ryu H, Kim J, Cho KS (2011) Rhizoremediation of diesel-contaminated soil using the plant growth-promoting rhizobacterium Gordonia sp. S2RP-17. Biodegradation 22(3):593–601. doi:10.1007/s10532-010-9432-2
Hundt K, Wagner M, Becher D, Hammer E, Schauer F (1998) Effect of selected environmental factors on degradation and mineralization of biaryl compounds by the bacterium Ralstonia pickettii in soil and compost. Chemosphere 36(10):2321–2335
Jigami Y, Kawasaki Y, Omori T, Minoda Y (1979) Coexistence of different pathways in the metabolism of n-propylbenzene by Pseudomonas sp. Appl Environ Microbiol 38(5):783–788
Jussila MM, Jurgens G, Lindstrom K, Suominen L (2006) Genetic diversity of culturable bacteria in oil-contaminated rhizosphere of Galega orientalis. Environ Pollut 139(2):244–257. doi:10.1016/j.envpol.2005.05.013
Kästner M, Breuer-Jammali M, Mahro B (1994) Enumeration and characterization of the soil microflora from hydrocarbon-contaminated soil sites able to mineralize polycyclic aromatic hydrocarbons (PAH). Appl Microbiol Biotechnol 41(2):267–273
Khorasani AC, Mashreghi M, Yaghmaei S (2013) Study on biodegradation of Mazut by newly isolated strain Enterobacter cloacae BBRC10061: improving and kinetic investigation. Iran J Environ Health Sci Eng 10(1):2–9. doi:10.1186/1735-2746-10-2
Kim D, Kim YS, Kim SK, Kim SW, Zylstra GJ, Kim YM, Kim E (2002) Monocyclic aromatic hydrocarbon degradation by Rhodococcus sp. strain DK17. Appl Environ Microbiol 68(7):3270–3278. doi:10.1128/aem. 68.7.3270-3278.2002
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721. doi:10.1099/ijs. 0.038075-0
Koma D, Sakashita Y, Kubota K, Fujii Y, Hasumi F, Chung SY, Kubo M (2005) Degradation pathways of cyclic alkanes in Rhodococcus sp NDKK48. Appl Microbiol Biotechnol 66(1):92–99. doi:10.1007/s00253-004-1623-5
Komukai-Nakamura S, Sugiura K, Yamauchi-Inomata Y, Toki H, Venkateswaran K, Yamamoto S, Tanaka H, Harayama S (1996) Construction of bacterial consortia that degrade Arabian light crude oil. J Ferment Bioeng 82(6):570–574. doi:10.1016/s0922-338x(97)81254-8
Kuiper I, Lagendijk EL, Bloemberg GV, Lugtenberg BJ (2004) Rhizoremediation: a beneficial plant-microbe interaction. Mol Plant Microbe Interact 17(1):6–15. doi:10.1094/MPMI.2004.17.1.6
Kummer C, Schumann P, Stackebrandt E (1999) Gordonia alkanivorans sp. nov., isolated from tar-contaminated soil. Int J Syst Bacteriol 49 Pt 4:1513-22
Lin C-L, Shen F-T, Tan C-C, Huang C-C, Chen B-Y, Arun AB, Young C-C (2012) Characterization of Gordonia sp. strain CC-NAPH129-6 capable of naphthalene degradation. Microbiol Res 167(7):395–404. doi:10.1016/j.micres.2011.12.002
Lo Piccolo L, De Pasquale C, Fodale R, Puglia AM, Quatrini P (2011) Involvement of an alkane hydroxylase system of Gordonia sp strain SoCg in degradation of solid n-alkanes. Appl Environ Microbiol 77(4):1204–1213. doi:10.1128/aem. 02180-10
Maila MP, Randima P, Cloete TE (2005) Multispecies and monoculture rhizoremediation of polycyclic aromatic hydrocarbons (PAHs) from the soil. Int J Phytoremediat 7(2):87–98. doi:10.1080/16226510590950397
McKenna EJ, Kallio RE (1971) Microbial metabolism of the isoprenoid alkane pristane. Proc Natl Acad Sci U S A 68(7):1552–1554
Mikolasch A, Hammer E, Schauer F (2003) Synthesis of imidazol-2-yl amino acids by using cells from alkane-oxidizing bacteria. Appl Environ Microbiol 69(3):1670–1679. doi:10.1128/aem. 69.3.1670-1679.2003
Morgan P, Watkinson RJ (1994) Biodegradation of components of petroleum. In: Ratledge C (ed) Biochemistry of microbial degradation. Kluwer, Dordrecht, pp 1–31
Nhi-Cong LT, Mikolasch A, Klenk H-P, Schauer F (2009) Degradation of the multiple branched alkane 2,6,10,14-tetramethyl-pentadecane (pristane) in Rhodococcus ruber and Mycobacterium neoaurum. Int Biodeterior Biodegrad 63(2):201–207. doi:10.1016/j.ibiod.2008.09.002
Nhi-Cong LT, Mikolasch A, Awe S, Sheikhany H, Klenk H-P, Schauer F (2010) Oxidation of aliphatic, branched chain, and aromatic hydrocarbons by Nocardia cyriacigeorgica isolated from oil-polluted sand samples collected in the Saudi Arabian Desert. J Basic Microbiol 50(3):241–253. doi:10.1002/jobm.200900358
Nicdao MAC, Rivera WL (2012) Two strains of Gordonia terrae isolated from used engine oil contaminated soil utilize short- to long-chain n-alkanes. Phil Sci Lett 5(2):199–208
Owsianiak M, Szulc A, Chrzanowski L, Cyplik P, Bogacki M, Olejnik-Schmidt AK, Heipieper HJ (2009) Biodegradation and surfactant-mediated biodegradation of diesel fuel by 218 microbial consortia are not correlated to cell surface hydrophobicity. Appl Microbiol Biotechnol 84(3):545–553. doi:10.1007/s00253-009-2040-6
Perry JJ (1977) Microbial metabolism of cyclic hydrocarbons and related compounds. Crit Rev Microbiol 5(4):387–412. doi:10.3109/10408417709102811
Poole PR, Whitaker G (1997) Biotransformation of 6-pentyl-2-pyrone by Botrytis cinerea in liquid cultures. J Agric Food Chem 45(1):249–252. doi:10.1021/jf9603644
Rapp P, Gabriel-Jürgens LH (2003) Degradation of alkanes and highly chlorinated benzenes, and production of biosurfactants, by a psychrophilic Rhodococcus sp. and genetic characterization of its chlorobenzene dioxygenase. Microbiology 149(Pt 10):2879–2890
Ratledge C (1978) Degradation of aliphatic hydrocarbons. In: Watkinson RJ (ed) Developments in biodegradation of hydrocarbons, vol 1. Applied Science, London, pp 1–46
Rojo F (2009) Degradation of alkanes by bacteria. Environ Microbiol 11(10):2477–2490. doi:10.1111/j.1462-2920.2009.01948.x
Rontani JF, Bonin P (1992) Utilization of n-alkyl-substituted cyclohexanes by a marine Alcaligenes. Chemosphere 24(10):1441–1446. doi:10.1016/0045-6535(92)90266-t
Sariasla FS, Harper DB, Higgins IJ (1974) Microbial degradation of hydrocarbons catabolism of 1-phenyl-alkanes by Nocardia salmonicolor. Biochem J 140(1):31–45
Schumann P, Maier T (2014) MALDI-TOF mass spectrometry applied to classification and identification of bacteria. Method Microbiol. doi:10.1016/bs.mim.2014.06.002
Shen F-T, Young L-S, Hsieh M-F, Lin S-Y, Young C-C (2010) Molecular detection and phylogenetic analysis of the alkane 1-monooxygenase gene from Gordonia spp. Syst Appl Microbiol 33(2):53–59. doi:10.1016/j.syapm.2009.11.003
Singh SN, Kumari B, Mishra S (2012) Microbial degradation of alkanes. In: Singh SN (ed) Microbial degradation of xenobiotics, environmental science and engineering. Springer-Verlag, Berlin, Heidelberg, pp 439–469
Smith DI, Callely AG (1975) The microbial degradation of cyclohexane carboxylic acid. J Gen Microbiol 91(NOV):210–212
Smith MR, Ratledge C (1989) Catabolism of alkylbenzenes by Pseudomonas sp. NCIB 10643. Appl Microbiol Biotechnol 32(1):68–75
Solano-Serena F, Marchal R, Vandecasteele J-P (2008) Biodegradation of aliphatic and alicyclic hydrocarbons. In: Vandecasteele J-P (ed) Petroleum microbiology: concepts, environmental implications, industrial applications, vol 1. Editions Technip, Paris, pp 173–240
Soleimani M, Afyuni M, Hajabbasi MA, Nourbakhsh F, Sabzalian MR, Christensen JH (2010) Phytoremediation of an aged petroleum contaminated soil using endophyte infected and non-infected grasses. Chemosphere 81(9):1084–1090. doi:10.1016/j.chemosphere.2010.09.034
Song HG, Wang XP, Bartha R (1990) Bioremediation potential of terrestrial fuel spills. Appl Environ Microbiol 56(3):652–656
Song X, Xu Y, Li G, Zhang Y, Huang T, Hu Z (2011) Isolation, characterization of Rhodococcus sp. P14 capable of degrading high-molecular-weight polycyclic aromatic hydrocarbons and aliphatic hydrocarbons. Mar Pollut Bull 62(10):2122–2128. doi:10.1016/j.marpolbul.2011.07.013
Stead DE, Sellwood JE, Wilson J, Viney I (1992) Evaluation of a commercial microbial identification system based on fatty acid profiles for rapid, accurate identification of plant pathogenic bacteria. J Appl Bacteriol 72:315–321. doi:10.1111/j.1365-2672.1992.tb01841.x
Stope MB, Becher D, Hammer E, Schauer F (2002) Cometabolic ring fission of dibenzofuran by Gram-negative and Gram-positive biphenyl-utilizing bacteria. Appl Microbiol Biotechnol 59(1):62–67. doi:10.1007/s00253-0002-0979-7
Suslow TV, Schroth MN, Isaka M (1982) Application of a rapid method for Gram differentiation of plant pathogenic and saprophytic bacteria without staining. Phytopathology 72:917–918. doi:10.1094/Phyto-77-917
Takeuchi M, Hatano K (1998) Gordonia rhizosphera sp. nov, isolated from the mangrove rhizosphere. Int J Syst Bacteriol 48:907–912
Táncsics A, Benedek T, Farkas M, Máthé I, Márialigeti K, Szoboszlay S, Kukolya J, Kriszt B (2014) Sequence analysis of 16S rRNA, gyrB and catA genes and DNA–DNA hybridization reveal that Rhodococcus jialingiae is a later synonym of Rhodococcus qingshengii. Int J Syst Evol Microbiol 64:298–301
Tjessem K, Aaberg A (1983) Photochemical transformation and degradation of petroleum residues in the marine environment. Chemosphere 12(11–1):1373–1394. doi:10.1016/0045-6535(83)90070-x
Upadhyay SK, Singh DP, Saikia R (2009) Genetic diversity of plant growth promoting rhizobacteria isolated from rhizospheric soil of wheat under saline condition. Curr Microbiol 59(5):489–496. doi:10.1007/s00284-009-9464-1
van Beilen JB, Li Z, Duetz WA, Smits THM, Witholt B (2003) Diversity of alkane hydroxylase systems in the environment. Oil Gas Sci Technol 58(4):427–440. doi:10.2516/ogst:2003026
Waldau D, Methling K, Mikolasch A, Schauer F (2009) Characterization of new oxidation products of 9H-carbazole and structure related compounds by biphenyl-utilizing bacteria. Appl Microbiol Biotechnol 81(6):1023–1031. doi:10.1007/s00253-008-1723-8
Wang XP, Bartha R (1990) Effects of bioremediation on residues, activity and toxicity in soil contaminated by fuel spills. Soil Biol Biochem 22(4):501–505. doi:10.1016/0038-0717(90)90185-3
Warhurst AM, Fewson CA (1994) Biotransformations catalyzed by the genus Rhodococcus. Crit Rev Biotechnol 14(1):29–73. doi:10.3109/07388559409079833
Watkinson RJ, Morgan P (1990) Physiology of aliphatic hydrocarbon-degrading microorganisms. Biodegradation 1(2–3):79–92
Webley DM, Duff RB, Farmer VC (1956) Evidence for β-oxidation in the metabolism of saturated aliphatic hydrocarbons by soil species of Nocardia. Nature 178(4548):1467–1468. doi:10.1038/1781467b0
Werle E, Schneider C, Renner M, Volker M, Fiehn W (1994) Convenient single-step, one tube purification of PCR products for direct sequencing. Nucleic Acids Res 22(20):4354–4355
White GF, Russell NJ (1994) Biodegradation of anionic surfactants and related molecules. In: Ratledge C (ed) Biochemistry of microbial degradation. Kluwer, Dordrecht, pp 143–177
Whyte LG, Hawari J, Zhou E, Bourbonniere L, Inniss WE, Greer CW (1998) Biodegradation of variable-chain-length alkanes at low temperatures by a psychrotrophic Rhodococcus sp. Appl Environ Microbiol 64(7):2578–2584
Widdel F, Musat F (2010) Diversity and common principles in enzymatic activation of hydrocarbons. In: Timmis KN (ed) Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, Heidelberg, pp 981–1009
Young CC, Lin TC, Yeh MS, Shen FT, Chang JS (2005) Identification and kinetic characteristics of an indigenous diesel-degrading Gordonia alkanivorans strain. World J Microbiol Biotechnol 21(8–9):1409–1414. doi:10.1007/s11274-005-5742-7
Acknowledgments
We thank R. Jack (Institute of Immunology, University of Greifswald) for help in preparing the manuscript. We thank Central Asian Biodiversity Network (CABNET), in particular the project manager Michael Manthey (Institute of Botany and Landscape Ecology, University of Greifswald), for the opportunity to establish active contacts between scientists from the Al-Farabi Kazakh National University and the University of Greifswald. Financial support from Deutscher Akademischer Austauschdienst (DAAD) (project code 50754935, project title “CABNET-Central Asian Biodiversity Network”) is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 590 kb)
Rights and permissions
About this article
Cite this article
Mikolasch, A., Omirbekova, A., Schumann, P. et al. Enrichment of aliphatic, alicyclic and aromatic acids by oil-degrading bacteria isolated from the rhizosphere of plants growing in oil-contaminated soil from Kazakhstan. Appl Microbiol Biotechnol 99, 4071–4084 (2015). https://doi.org/10.1007/s00253-014-6320-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-6320-4