Abstract
Aims/hypothesis
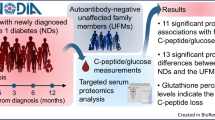
We sought to identify minimal sets of serum peptide signatures as markers for islet autoimmunity and predictors of progression rates to clinical type 1 diabetes in a case–control study.
Methods
A double cross-validation approach was applied to first prioritise peptides from a shotgun proteomic approach in 45 islet autoantibody-positive and -negative children from the BABYDIAB/BABYDIET birth cohorts. Targeted proteomics for 82 discriminating peptides were then applied to samples from another 140 children from these cohorts.
Results
A total of 41 peptides (26 proteins) enriched for the functional category lipid metabolism were significantly different between islet autoantibody-positive and autoantibody-negative children. Two peptides (from apolipoprotein M and apolipoprotein C-IV) were sufficient to discriminate autoantibody-positive from autoantibody-negative children. Hepatocyte growth factor activator, complement factor H, ceruloplasmin and age predicted progression time to type 1 diabetes with a significant improvement compared with age alone.
Conclusion/interpretation
Distinct peptide signatures indicate islet autoimmunity prior to the clinical manifestation of type 1 diabetes and enable refined staging of the presymptomatic disease period.
Similar content being viewed by others
Introduction
The development of type 1 diabetes includes an asymptomatic period of autoimmunity identified by the presence of islet autoantibodies, with subsequent progression to dysglycaemia and clinical diabetes [1]. While the development of islet autoantibodies is most prominent around 1–2 years of age [2–4], the incidence of clinical diabetes appears to be relatively constant in multiple islet autoantibody-positive children and adolescents [5]. Biomarkers and genetics that are associated with islet autoimmunity are of interest for elucidating pathogenesis, and biomarkers that predict the rate of progression [6–10] may improve staging the presymptomatic disease period of type 1 diabetes.
Proteomics has been used to identify biomarkers in diverse diseases such as cardiovascular diseases [11], prostrate and other cancers [12, 13], Parkinson’s disease [14] and metabolic disorders [11, 15]. In type 1 diabetes, previous proteomic biomarker screening studies have compared patients with type 1 diabetes to autoantibody-negative control participants [16–18] and identified protein signatures correlated with clinical disease. A recent longitudinal study in Finland compared islet autoantibody-positive children with autoantibody-negative children, and identified a protein signature that distinguished between healthy children and those with autoimmunity [19].
Here, we applied proteomics to our cohorts of children followed from birth to islet autoimmunity and clinical diabetes in order to search for signatures associated with islet autoimmunity, and which could help predict the progression rate to clinical diabetes in multiple autoantibody-positive children.
Methods
This study was performed using sera from children participating in either the BABYDIAB [20] or BABYDIET [21] studies. These birth cohort studies enrolled children with a family history of type 1 diabetes and are prospectively monitoring the natural history of islet autoimmunity and type 1 diabetes. Together, they have enrolled 2441 children [20, 21]. By November 2014, 124 children had developed multiple islet autoantibodies and 82 of these children had progressed to clinical type 1 diabetes [22].
Islet autoantibodies were measured using radiobinding assays as previously described [8, 20]. The antibody assays were evaluated in the Diabetes Autoantibody Standardization Program (Laboratory 121) [23–25]. Diabetes was diagnosed according to the ADA Expert Committee criteria [26]. Both studies were approved by the ethics committee of Bavaria, Germany (Bayerische Landesärztekammer No. 95357 and Ludwig-Maximilians University No. 329/00, respectively), and adhered to the principles of the Declaration of Helsinki.
Sample selection and study design
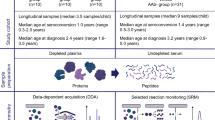
The analysis was performed in two phases: a peptide-selection phase in which shotgun proteomics was performed to identify peptides of potential interest, which were then measured by targeted proteomics in a second application phase (Fig. 1 and electronic supplementary material [ESM] Fig.1). For the selection phase, we applied shotgun proteomics to samples from children who developed islet autoantibodies and progressed to clinical diabetes within 3.5 years (‘rapid’ progression: 15 children; median follow-up from seroconversion 1.9 years, interquartile range [IQR] 1.0–2.9 years, range 0.5–3.3 years) or ≥9.5 years (‘slow’ progression: 15 children; median follow-up from seroconversion 14.5 years, IQR 12.9–15.5 years, range 9.5–17.4 years), and from 15 children who remained islet autoantibody-negative (median follow-up from birth 15.9 years, IQR 14.2–17.4 years, range 5.9–21.7 years) matched for sex and age (Fig. 1). Two sample times were separately analysed. Specifically, one sample from each child was obtained shortly after seroconversion to the first islet autoantibody (median 0.8 years, IQR 0.3–1.4 years; sample set 1) or at the corresponding age in islet autoantibody-negative children, while the other sample was obtained at a later time (median 1.2 years after the first sample, IQR 0.8–2.9 years; sample set 2). Four children were excluded from sample set 2 in the selection phase because they had already progressed to overt diabetes by the time the second sample had been collected after seroconversion.
For the application phase, we randomly selected 70 of the remaining children who developed islet autoantibodies (median age 3.2 years, median follow-up time 12.8 years, IQR 9.6–16.6 years) and 70 sex- and age-matched islet autoantibody-negative children (median age 3.1 years, median follow-up time 10.8 years, IQR 7.2–14.4 years) (Fig. 1).
We performed targeted proteomics on the peptides that discriminated between groups in the selection phase (see detailed description below). Samples from the 70 islet autoantibody-positive children were obtained shortly after seroconversion (median 1.0 years, IQR 0.5–1.3 years; Fig. 1) and 60 children were multiple islet autoantibody-positive at the time of proteomics measurement.
Sample preparation for MS
Plasma samples were depleted from highly abundant proteins and proteolysed with trypsin as previously described [27]. All samples were randomly distributed into one of three batches for processing, and the experimenters were blinded to the sample-group allocation during the experiment. For quality control of depletion, digestion and MS measurements, each sample was spiked with ribulose-1,5-bisphosphate carboxylase oxygenase (Sigma Aldrich, Taufkirchen, Germany) at a final amount of 50 fmol in each 10 μl serum sample. After digestion, samples were stored at −80°C until further use.
Non-targeted liquid chromatography tandem MS (LC-MS/MS) and label-free quantification
LC-MS/MS analyses were performed as previously described [28] on an LTQ-Orbitrap XL instrument (Thermo Fisher Scientific, Dreieich, Germany) operated with an RSLC system (Ultimate 3000, Thermo Fisher Scientific). The RAW files (Thermo Fisher Scientific) were analysed using the Progenesis LC-MS software (version 4.0; Nonlinear Dynamics, Waters, Eschborn, Germany), as previously described [27, 29].
Targeted LC-MS/MS using selected reaction monitoring (SRM)
Skyline software (MacCoss Lab Software, Seattle, WA, USA) was used to create the SRM assays [30]. We developed and optimised an SRM assay if at least one peptide per protein satisfied the quality criteria defined using the AuDIT algorithm [31] for reproducible and reliable SRM measurement. Isotope-labelled, synthetic peptides (heavy peptides; PEPotec; Thermo Fisher Scientific, Ulm, Germany) were used as internal controls for correct signal integration and relative quantification. The heavy peptide mix was added to the digested sample before the MS measurement.
SRM-MS analyses were performed on a Tempo Nano MDLC system (Eksigent Technologies, Dublin, OH, USA) coupled online to a triple quadrupole QTrap4000 (AB SCIEX, Framingham, MA, USA) MS equipped with a nanospray ion source [27]. During the MS measurements, the preselected proteotypic peptides were fragmented and the areas under the chromatographic curves of the resulting transitions formed the basis of the SRM quantifications.
Processing of SRM data
SRM data were processed using the Skyline software as previously described [15]. Briefly, after manual quality control, heavy to light peptide ratios were calculated on fragment levels, log2 transformed and corrected for batch effects by linear regression, followed by averaging fragment values to peptides. The peptide values were normalised against control protein peptides and are referred to as adjusted intensities. Peptides with unreliable signals (>20% of measurements below the limits of quantification per peptide) were removed, resulting in robust SRM assays for 82 peptides covering 50 proteins (ESM Table 1).
Statistical analysis in the selection phase
In the selection phase, using a univariate non-parametric test (Wilcoxon rank-sum test), we assessed group differences in both sample sets (one collected shortly after seroconversion and one collected at a later time point) between: (1) islet autoantibody-positive vs autoantibody-negative children; (2) autoantibody-negative children vs slow progressors; (3) autoantibody-negative children vs rapid progressors; and (4) slow vs rapid progressors. Multiple hypothesis testing was corrected for by controlling the false discovery rate (FDR) at 0.05.
A double cross-validation (dCV) approach was then used to identify multivariable predictive protein and peptide signatures for the same eight comparisons (two sample sets and four group comparisons each). This approach selected a minimal combination of peptides that provided high discriminative accuracy, and estimated an unbiased, non-over-fitted AUC [32]. A detailed explanation of the approach and the parameter settings used in our study can be found in the ESM Method.
Peptides occurring with at least 75% selection frequency in at least one of the eight comparisons were compiled into a candidate ‘selection’ list. To maximise our coverage, this list was extended by 14 peptides that were reported in a recent proteomics study [17].
Statistical analysis in the application phase
In the application phase, we tested for differences in peptide levels between islet autoantibody-positive and autoantibody-negative children using Wilcoxon rank-sum tests. To model the time from seroconversion to type 1 diabetes, we fitted univariate Cox regression models within the islet autoantibody-positive samples. Multiple hypothesis testing was corrected for by controlling the FDR at 0.05. Highly correlated peptides were identified using Pearson’s correlation coefficient.
We again applied the dCV algorithm to find multivariable peptide signatures discriminating between islet autoantibody-positive and autoantibody-negative samples. A modified version of this algorithm that used Cox models instead of classification models was then applied to identify a predictive signature of progression time within the autoantibody-positive children. For the dCV analyses in the application phase, we also included age as an explanatory variable. Details on the dCV approach in the application phase can be found in the ESM Method.
Peptides with a selection frequency of at least 50% were used to fit a final Cox model, yielding progression time risk scores for each autoantibody-positive individual in the application set. These scores were divided into low-, medium- and high-risk tertiles. Differences in the survival curves between the tertiles were assessed using logrank tests. In order to investigate the improvement in discrimination conferred by the selected peptides in addition to age, a Cox model containing only age was compared with the combined model by ANOVA. In addition, the discrimination performance over time of the combined model and of age alone was evaluated using the survival AUC measure [33]. As an overall measure of discrimination, an integrated AUC was calculated.
All analyses were performed using R version 3.2.0 (www.r-project.org).
Enrichment analysis
GeneRanker software (Genomatix software suite V3.5; Genomatix, Munich, Germany) was used to evaluate protein enrichment. Gene symbols for the respective proteins were used as identifiers. Gene ontology enrichment was calculated by comparing all significantly different proteins identified in the application phase as discriminating between islet autoantibody-positive and autoantibody-negative children against all proteins identified in plasma in the discovery phase. Redundancies in enriched terms for biological processes were curated manually.
Results
Shotgun proteomics identified tryptic peptides, which discriminated between autoantibody statuses and progression rates
Shotgun proteomics of serum samples from the selection group resulted in the quantification of 2021 tryptic peptides (covering 204 proteins) in the first sample set and 2996 tryptic peptides (243 proteins) in the second sample set. A total of 215 peptides (covering 106 proteins) were selected by the dCV approach for discrimination in at least one between-group comparison (islet autoantibody-positive vs autoantibody-negative; slow vs rapid; autoantibody-negative vs slow; and autoantibody-negative vs rapid). Of these, 169 peptides overlapped between the first and second sample sets and were evaluated for SRM development.
Robust SRM assays were developed for 82 peptides (covering 50 proteins; ESM Table 1). These included 14 peptides that were added from a previous study [17] but were not selected as significant in the selection phase of this study (ESM Table 1).
Application phase: targeted proteomic analyses for discriminating between islet autoantibody-positive and autoantibody-negative children
In univariate analysis, the abundance of 26 proteins (represented by 41 peptides) differed significantly between autoantibody-positive and autoantibody-negative children (Table 1; ESM Fig. 2). Eight of those proteins (represented by 14 peptides) overlapped with findings from previous studies [16–19] (Table 1). This included four of the 14 peptides that were tested in our study because they had been identified in a previous study [17] (Table 1). Pearson’s correlation test revealed several correlated peptides. As expected, the highest correlations were observed for peptides belonging to the same protein, indicating a high reliability of SRM measurements (ESM Fig. 3). Peptides representing proteins belonging to the same protein family, such as apolipoproteins (APOs), also showed highly correlated abundance patterns (ESM Fig. 3). Gene ontology enrichment analysis recovered a significant accumulation of differentially abundant proteins in terms associated with lipid metabolic processes and homeostasis, indicative of changes in lipid metabolism (ESM Table 2).
The multivariable dCV method selected two peptides, SLTSCLDSK from APOM and ELLETVVNR from APOC4, to discriminate between islet autoantibody-positive and autoantibody-negative children (Fig. 2, Table 2) and yielded an unbiased median AUC of 0.77 (IQR 0.75–0.78). Using the logistic regression coefficients (Table 2) as weights, we calculated a combined risk score to discriminate between islet autoantibody-positive and autoantibody-negative children based solely on these two peptides (Fig. 2). The AUC of 0.83 for this combined model was significantly higher than that for APOM alone (AUC 0.75) and for APOC4 alone (AUC 0.74) at p = 2.5 × 10−5.
Quantitative differences of the best discriminating peptides in islet autoantibody-positive (AB+) and autoantibody-negative (AB−) children. Quantifications are based on SRM measurements in the application sample set. (a) Adjusted peptide (SLTSCLDSK) intensities of APOM (Wilcoxon test, p = 2.5 × 10−5) in arbitrary units (AU). (b) Adjusted peptide (ELLETVVNR) intensities of APOC4 (p = 2.5 × 10−5). (c) Risk score for the final model. The risk score was calculated using a logistic regression model with the selected peptides using the weights shown in Table 2
Targeted proteomics to predict disease progression
There were no significant univariate associations of individual peptides with progression time.
When we applied the survival dCV approach, we found that three peptides (representing hepatocyte growth factor activator [HGFAC], complement factor [CF]H and ceruloplasmin [CP]) and age at measurement were predictive covariates for progression time (Fig. 3a–c, Table 2). The median survival AUC was 0.72 (IQR 0.69–0.75). In order to investigate the improvement in discrimination conferred by the three peptides in addition to age, we compared the AUC of the combined model with that of age alone (ESM Fig. 4). The combined model displayed a significant improvement in discrimination (p = 0.001), mainly due to an improvement after 4 years of follow-up. Importantly, the abundance levels of most peptides, including the three peptides predictive for progression time, were not correlated with age (ESM Fig. 5). Only the levels of both peptides representing carnosine dipeptidase 1 significantly increased with age (ESM Fig. 5) but these peptides were not, however, selected by the dCV for either progression rates or autoimmunity status.
Progression time analysis. Adjusted intensities of the selected peptides of (a) HGFAC (VANYVDWINDR), (b) CFH (SSIDIENGFISESQYTYALK) and (c) CP (HYYIGIIETTWDYASDHGEK) in arbitrary units (AU) and the corresponding time from seroconversion to type 1 diabetes in the application cohort. (d) Kaplan–Meier curves of the high-, medium- and low-risk score groups (defined by age, HGFAC, CFH and CP) for the time from seroconversion to type 1 diabetes. Blue line, low-risk group; grey line, medium-risk group; red line, high-risk group; dotted lines, CIs; dashed line, 5 year interval. The low- and high-risk survival curves were significantly different (p = 1.6 × 10−5). The numbers of children remaining at risk at a given time are shown below the time axis. SC, seroconversion; T1D, type 1 diabetes
Using these peptides and age, we calculated risk scores by including the weights from Table 2 in a multivariable Cox model, and separated the children into tertiles of high, medium and low risk (Fig. 3d; ESM Table 3). Children in the low-risk group progressed to type 1 diabetes with a probability of <10% within 5 years after seroconversion (95% CI 2.2%, 29%). The corresponding rate in the high risk group was 78% (95% CI 60%, 92%) (Fig. 3d). As expected, children in the high-risk group were younger; however, they did not differ from the other risk groups in islet autoantibody status or HLA genotype (ESM Table 3).
Discussion
Using a proteomics strategy, we determined the protein expression profiles of 185 children from the BABYDIAB/BABYDIET birth cohorts with high genetic risk for type 1 diabetes. We found that 26 proteins, represented by 41 peptides, could discriminate between islet autoantibody-positive and autoantibody-negative children. The 26 proteins were enriched for pathways involved in lipid-associated metabolic processes and homeostasis, suggesting that changes in lipid metabolism occur early in the autoimmunity process. We also identified a proteomic signature that, together with age, was able to discriminate fast and slow progression to clinical diabetes in islet autoantibody-positive children.
Previous studies have used LC-MS/MS-based proteomics approaches and applied extensive prefractionation techniques on pooled samples [17, 18], followed by applying selected candidate proteins using ELISA [18], LC-SRM-MS [18] or other methods [19]. We designed our study in two phases, capitalising on the high analytical depth of a shotgun proteomics approach for selecting interesting peptides followed by an application using sensitive targeted proteomics specifically developed for the subset of potentially relevant peptides. The technical advantages of the targeted proteomics approach include high accuracy and robustness of quantifications, and that all peptides are consistently measured across all LC-MS runs, thus avoiding the occurrence of missing values.
Consistent with previous studies in children with overt type 1 diabetes, we found lower levels of APOA4 [19], APOC4 [19], CF3 [17, 18], CF4 [17–19], clusterin [16, 17], kininogen [17] and transthyretin [17] in children with islet autoantibodies. We also found lower levels of CF9 in autoantibody-positive children, while others [19] have reported slightly increased levels. The peptide of APOM that was selected for discrimination in the risk score has not been identified in previous studies. In addition, we identified changes in the levels of 18 proteins (represented by 27 peptides) that have not been previously described.
In order to prioritise the peptide signatures, we used the dCV method for feature selection. This method aims to derive a minimal, predictive combination of peptides, and to estimate the predictive power within a dataset in an unbiased fashion, without substantial overfitting effects [32]. Two peptides, one from APOM and one from APOC4, were deemed to be sufficient for between-group discrimination with a median AUC of 0.83. Both peptides were also among the top hits for discrimination in the univariate analysis. APOM levels were higher and APOC4 levels lower in the children with islet autoantibodies in our study. APOM is a member of the lipocalin protein family involved in lipid transport [34]. Polymorphisms in the promoter region of APOM that increase promoter activity have also been reported to increase susceptibility to the development of type 1 diabetes in two different cohorts [35]. Because the APOM gene is regulated by hepatocyte nuclear factor 1α (HNF1A), APOM is also considered to be a marker of HNF1A-dependent MODY. However, APOM levels have been found to be significantly lower in individuals with MODY than in those with type 1 diabetes [36].
APOC4, the other major marker for discriminating between islet autoantibody-positive and autoantibody-negative children in this study, is also a member of the APO family. The lower levels of APOC4 in autoantibody-positive children confirm previous findings reported in the Finnish Type 1 Diabetes Prediction and Prevention (DIPP) study [19], in which APOC4 levels were decreased even before seroconversion in children who eventually progressed to type 1 diabetes. Lower APOA1 levels have been reported to be associated with viral infections [37], and Moulder et al have suggested an association between viral infections and the development of type 1 diabetes [19]. So far, to the best of our knowledge, APOC4 has not been described to play a role in the immune response. However, recent studies have discussed APOs such as APOM [38] in the context of autoimmunity [38–40], and future studies might unravel as-yet unidentified roles for APOC4.
The combined discriminative power of the candidate proteins APOM and APOC4 (median AUC 0.83) is comparable with the results reported for APOC4 and afamin (AUC 0.85) in the DIPP study [19].
Another aim was to explore whether proteomic signatures could predict the progression time to type 1 diabetes in children with islet autoantibodies. We identified a set of three peptides representing three proteins, CFH, HGFAC and CP, in addition to age, as predictive covariates for progression time with a median survival AUC of 0.72. Predictions including these peptides were slightly but significantly superior to those using age alone. Higher levels of CFH and HGFAC and lower levels of CP in combination with young age were associated with faster progression in later follow-up. CFH, HGFAC and CP have previously been discussed in relation to insulin resistance [41], type 1 diabetes [17] and type 2 diabetes [42, 43], respectively.
The strengths and novelties of our study included the multivariate statistical approach for extracting relevant peptide signatures, minimising false-positive associations; the exclusive investigation of samples from patients close to seroconversion without overt diabetes, thus reducing the confounding effect of hyperglycaemia on proteomic signatures; and the large cohort of children with islet autoimmunity. A limitation of our study is that we did not validate our signature of progression rate in a separate cohort. Other limitations include the lack of repeated longitudinal measurements and the relatively small contribution of the peptide signature to the progression risk score, as compared with age alone.
In conclusion, we found that serum proteomics signatures of islet autoantibody-positive children close to the date of seroconversion were dominated by proteins involved in lipid metabolism. Some of these protein markers have been previously identified in studies of patients with overt diabetes, and the changes in their levels close to the onset of autoimmunity suggest they are early markers. In addition, the peptide signatures significantly improved the categorisation of islet autoantibody-positive children into high- or low-risk groups for rapid progression to type 1 diabetes over age alone.
Abbreviations
- APO:
-
Apolipoprotein
- CF:
-
Complement factor
- CP:
-
Ceruloplasmin
- dCV:
-
Double cross-validation
- DIPP study:
-
Type 1 Diabetes Prediction and Prevention study
- FDR:
-
False discovery rate
- HGFAC:
-
Hepatocyte growth factor activator
- HNF1A:
-
Hepatocyte nuclear factor 1α
- IQR:
-
Interquartile range
- LC-MS/MS:
-
Liquid chromatography tandem MS
- SRM:
-
Selected reaction monitoring
References
Insel RA, Dunne JL, Atkinson MA et al (2015) Staging presymptomatic type 1 diabetes: a scientific statement of JDRF, the Endocrine Society, and the American Diabetes Association. Diabetes Care 38:1964–1974
Ziegler AG, Bonifacio E (2012) Age-related islet autoantibody incidence in offspring of patients with type 1 diabetes. Diabetologia 55:1937–1943
Parikka V, Nanto-Salonen K, Saarinen M et al (2012) Early seroconversion and rapidly increasing autoantibody concentrations predict prepubertal manifestation of type 1 diabetes in children at genetic risk. Diabetologia 55:1926–1936
Krischer JP, Lynch KF, Schatz DA et al (2015) The 6 year incidence of diabetes-associated autoantibodies in genetically at-risk children: the TEDDY study. Diabetologia 58:980–987
Ziegler AG, Rewers M, Simell O et al (2013) Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. JAMA 309:2473–2479
Achenbach P, Bonifacio E, Williams AJ, Ziegler AG, Gale EA, Bingley PJ (2008) Autoantibodies to IA-2β improve diabetes risk assessment in high-risk relatives. Diabetologia 51:488–492
Achenbach P, Warncke K, Reiter J et al (2004) Stratification of type 1 diabetes risk on the basis of islet autoantibody characteristics. Diabetes 53:384–392
Achenbach P, Lampasona V, Landherr U et al (2009) Autoantibodies to zinc transporter 8 and SLC30A8 genotype stratify type 1 diabetes risk. Diabetologia 52:1881–1888
Achenbach P, Koczwara K, Knopff A, Naserke H, Ziegler AG, Bonifacio E (2004) Mature high-affinity immune responses to (pro)insulin anticipate the autoimmune cascade that leads to type 1 diabetes. J Clin Invest 114:589–597
Mayr A, Schlosser M, Grober N et al (2007) GAD autoantibody affinity and epitope specificity identify distinct immunization profiles in children at risk for type 1 diabetes. Diabetes 56:1527–1533
Yassine H, Borges CR, Schaab MR et al (2013) Mass spectrometric immunoassay and MRM as targeted MS-based quantitative approaches in biomarker development: potential applications to cardiovascular disease and diabetes. Proteomics Clin Appl 7:528–540
Pin E, Fredolini C, Petricoin EF 3rd (2013) The role of proteomics in prostate cancer research: biomarker discovery and validation. Clin Biochem 46:524–538
Chambers AG, Percy AJ, Simon R, Borchers CH (2014) MRM for the verification of cancer biomarker proteins: recent applications to human plasma and serum. Expert Rev Proteomics 11:137–148
Alberio T, Bucci EM, Natale M et al (2013) Parkinson’s disease plasma biomarkers: an automated literature analysis followed by experimental validation. J Proteomics 90:107–114
von Toerne C, Huth C, de Las Heras Gala T et al (2016) MASP1, THBS1, GPLD1 and ApoA-IV are novel biomarkers associated with prediabetes: the KORA F4 study. Diabetologia 59:1882–1892
Metz TO, Qian WJ, Jacobs JM et al (2008) Application of proteomics in the discovery of candidate protein biomarkers in a diabetes autoantibody standardization program sample subset. J Proteome Res 7:698–707
Zhang Q, Fillmore TL, Schepmoes AA et al (2013) Serum proteomics reveals systemic dysregulation of innate immunity in type 1 diabetes. J Exp Med 210:191–203
Zhi W, Sharma A, Purohit S et al (2011) Discovery and validation of serum protein changes in type 1 diabetes patients using high throughput two dimensional liquid chromatography-mass spectrometry and immunoassays. Mol Cell Proteomics 10:M111.012203
Moulder R, Bhosale SD, Erkkila T et al (2015) Serum proteomes distinguish children developing type 1 diabetes in a cohort with HLA-conferred susceptibility. Diabetes 64:2265–2278
Ziegler AG, Hummel M, Schenker M, Bonifacio E (1999) Autoantibody appearance and risk for development of childhood diabetes in offspring of parents with type 1 diabetes: the 2-year analysis of the German BABYDIAB Study. Diabetes 48:460–468
Hummel S, Pfluger M, Hummel M, Bonifacio E, Ziegler AG (2011) Primary dietary intervention study to reduce the risk of islet autoimmunity in children at increased risk for type 1 diabetes: the BABYDIET study. Diabetes Care 34:1301–1305
Giannopoulou EZ, Winkler C, Chmiel R et al (2015) Islet autoantibody phenotypes and incidence in children at increased risk for type 1 diabetes. Diabetologia 58:2317–2323
Torn C, Mueller PW, Schlosser M, Bonifacio E, Bingley PJ (2008) Diabetes Antibody Standardization Program: evaluation of assays for autoantibodies to glutamic acid decarboxylase and islet antigen-2. Diabetologia 51:846–852
Schlosser M, Mueller PW, Torn C, Bonifacio E, Bingley PJ (2010) Diabetes Antibody Standardization Program: evaluation of assays for insulin autoantibodies. Diabetologia 53:2611–2620
Lampasona V, Schlosser M, Mueller PW et al (2011) Diabetes antibody standardization program: first proficiency evaluation of assays for autoantibodies to zinc transporter 8. Clin Chem 57:1693–1702
Expert Committee on the Diagnosis and Classification of Diabetes Mellitus (2003) Report of the expert committee on the diagnosis and classification of diabetes mellitus. Diabetes Care 26(Suppl 1):S5–20
von Toerne C, Kahle M, Schafer A et al (2013) Apoe, Mbl2, and Psp plasma protein levels correlate with diabetic phenotype in NZO mice—an optimized rapid workflow for SRM-based quantification. J Proteome Res 12:1331–1343
Graessel A, Hauck SM, von Toerne C et al (2015) A combined omics approach to generate the surface atlas of human naive CD4+ T cells during early T-cell receptor activation. Mol Cell Proteomics 14:2085–2102
Hauck SM, Dietter J, Kramer RL et al (2010) Deciphering membrane-associated molecular processes in target tissue of autoimmune uveitis by label-free quantitative mass spectrometry. Mol Cell Proteomics 9:2292–2305
MacLean B, Tomazela DM, Shulman N et al (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26:966–968
Abbatiello SE, Mani DR, Keshishian H, Carr SA (2010) Automated detection of inaccurate and imprecise transitions in peptide quantification by multiple reaction monitoring mass spectrometry. Clin Chem 56:291–305
Laimighofer M, Krumsiek J, Buettner F, Theis FJ (2016) Unbiased prediction and feature selection in high-dimensional survival regression. J Comput Biol 23:279–290
Uno H, Cai T, Pencina MJ, D’Agostino RB, Wei LJ (2011) On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat Med 30:1105–1117
Xu N, Dahlback B (1999) A novel human apolipoprotein (apoM). J Biol Chem 274:31286–31290
Wu X, Niu N, Brismar K et al (2009) Apolipoprotein M promoter polymorphisms alter promoter activity and confer the susceptibility to the development of type 1 diabetes. Clin Biochem 42:17–21
Mughal SA, Park R, Nowak N et al (2013) Apolipoprotein M can discriminate HNF1A-MODY from type 1 diabetes. Diabet Med 30:246–250
Singh IP, Chopra AK, Coppenhaver DH, Ananatharamaiah GM, Baron S (1999) Lipoproteins account for part of the broad non-specific antiviral activity of human serum. Antiviral Res 42:211–218
Tsai HC, Han MH (2016) Sphingosine-1-phosphate (S1P) and S1P signaling pathway: therapeutic targets in autoimmunity and inflammation. Drugs 76:1067–1079
Ley K (2016) 2015 Russell Ross Memorial Lecture in Vascular Biology: protective autoimmunity in atherosclerosis. Arterioscler Thromb Vasc Biol 36:429–438
Black LL, Srivastava R, Schoeb TR, Moore RD, Barnes S, Kabarowski JH (2015) Cholesterol-independent suppression of lymphocyte activation, autoimmunity, and glomerulonephritis by apolipoprotein A-I in normocholesterolemic lupus-prone mice. J Immunol 195:4685–4698
Moreno-Navarrete JM, Martinez-Barricarte R, Catalan V et al (2010) Complement factor H is expressed in adipose tissue in association with insulin resistance. Diabetes 59:200–209
Cunningham J, Leffell M, Mearkle P, Harmatz P (1995) Elevated plasma ceruloplasmin in insulin-dependent diabetes mellitus: evidence for increased oxidative stress as a variable complication. Metabolism 44:996–999
Memisogullari R, Bakan E (2004) Levels of ceruloplasmin, transferrin, and lipid peroxidation in the serum of patients with type 2 diabetes mellitus. J Diabetes Complications 18:193–197
Acknowledgements
The authors thank S. Becker (Research Unit Protein Science, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany) for her excellent technical assistance.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Funding
This work was funded by JDRF grant 17-2012-617 and the iMED programme of the Helmholtz Association. The research programmes have also received funding from the EU’s Seventh Framework Program [FP7-Health-F5-2012] under grant agreement no. 305280 (MIMOmics). This work was further supported by grants from the German Federal Ministry of Education and Research to the German Center for Diabetes Research (DZD e.V.).
Duality of interest statement
The authors declare that there is no duality of interest associated with this manuscript.
Author contributions
CvT designed the study, performed and analysed the MS experiments, and wrote the manuscript. ML designed and performed statistical analyses and wrote the manuscript. PA performed sample selection and reviewed/edited the manuscript. AB performed statistical analyses and reviewed/edited the manuscript. TdlHG developed the procedure to process the SRM data. JK and FJT supervised the statistical analysis and reviewed/edited the manuscript. AGZ designed the study, supervised sample collection and selection, and wrote the manuscript. SMH designed the study, supervised the MS experiments and wrote the manuscript. CvT, ML and PA are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. All authors contributed to data interpretation, critically reviewed and edited the manuscript, and approved the version to be published.
Additional information
Christine von Toerne and Michael Laimighofer contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 2.15 mb)
Rights and permissions
About this article
Cite this article
von Toerne, C., Laimighofer, M., Achenbach, P. et al. Peptide serum markers in islet autoantibody-positive children. Diabetologia 60, 287–295 (2017). https://doi.org/10.1007/s00125-016-4150-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-016-4150-x