Abstract
Aims/hypothesis
Reduced oxidative capacity of the mitochondria in skeletal muscle has been suggested to contribute to insulin resistance and type 2 diabetes. Moreover, a set of genes influencing oxidative phosphorylation (OXPHOS) is downregulated in diabetic muscle. Here we studied whether genetic, epigenetic and non-genetic factors influence a component of the respiratory chain, COX7A1, previously shown to be downregulated in skeletal muscle from patients with type 2 diabetes. The specific aims were to: (1) evaluate the impact of genetic (single nucleotide polymorphisms [SNPs]), epigenetic (DNA methylation) and non-genetic (age) factors on the expression of COX7A1 in human skeletal muscle; and (2) investigate whether common variants in the COX7A1 gene are associated with increased risk of type 2 diabetes.
Methods
COX7A1 mRNA expression was analysed in muscle biopsies from young (n = 110) and elderly (n = 86) non-diabetic twins and related to measures of in vivo metabolism. Genetic variants (three SNPs) from the COX7A1 locus were genotyped in the twins and in two independent type 2 diabetes case–control cohorts (n = 1466 and 6380, respectively). DNA methylation of the COX7A1 promoter was analysed in a subset of twins (ten young, ten elderly) using bisulphite sequencing.
Results
While DNA methylation of the COX7A1 promoter was increased in muscle from elderly compared with young twins (19.9 ± 8.3% vs 1.8 ± 2.7%; p = 0.035), the opposite was found for COX7A1 mRNA expression (elderly 1.00 ± 0.05 vs young 1.68 ± 0.06; p = 0.0005). The heritability of COX7A1 expression was estimated to be 50% in young and 72% in elderly twins. One of the polymorphisms investigated, rs753420, influenced basal COX7A1 expression in muscle of young (p = 0.0001) but not of elderly twins. The transcript level of COX7A1 was associated with increased in vivo glucose uptake and \(\mathop V\limits^ \cdot {\text{O}}_{{\text{2max}}} \) (p = 0.009 and p = 0.001, respectively). We did not observe any genetic association between COX7A1 polymorphisms and type 2 diabetes after correcting for multiple testing.
Conclusions/interpretation
Our results provide further evidence for age as a factor influencing DNA methylation and expression of OXPHOS genes, and thereby in vivo metabolism.
Similar content being viewed by others
Introduction
Type 2 diabetes is characterised by insulin resistance in the liver and peripheral target tissues, hyperglycaemia and impaired pancreatic beta cell function. Reduced oxidative capacity of the mitochondria in skeletal muscle has been found to be associated with insulin resistance in type 2 diabetes [1]. In addition, a number of gene expression profiles have shown reduced levels of genes involved in oxidative phosphorylation (OXPHOS) and reduced levels of their transcriptional regulators, e.g. PGC-1α (also known as PPARGC1A) and PGC-1β (also known as PPARGC1B), in skeletal muscle from patients with type 2 diabetes [2–5]. Together, these reports propose that defects in the skeletal muscle respiratory chain may contribute to the pathogenesis of type 2 diabetes.
OXPHOS is a process where electrons are passed from NADH and FADH2 along a series of carrier molecules. In this process protons are pumped across the inner mitochondrial membrane to produce a proton gradient and in a final step this gradient is used to drive the production of ATP from ADP and phosphate. This process occurs in a system that consists of five multiprotein complexes with about 90 known protein subunits. While the majority of these subunits are encoded by the nuclear genome, 13 subunits are encoded by the 16.6 kb mitochondrial genome (mitochondrial DNA [mtDNA]).
COX7A1 is a nuclear encoded OXPHOS gene encoding polypeptide 1 of subunit 7A, a subunit of cytochrome c oxidase (COX), i.e. complex IV in the respiratory chain. COX catalyses the electron transfer from reduced cytochrome c to oxygen, producing H2O. This reaction is coupled with proton transfer from the matrix to the intermembrane space, thereby contributing to the energy stored in the electrochemical gradient. The catalytic core of COX is composed of mitochondrial encoded proteins whereas nuclear encoded proteins contribute to the structural and regulatory subunits of the complex. COX7A1 is located on chromosome 19q13.12 within a CpG dense region [6] where tissue-specific DNA methylation has been correlated with gene expression [7]. COX7A1 is only expressed in skeletal and heart muscle [8], and is significantly downregulated in diabetic muscle [3].
Type 2 diabetes develops as a conspiracy between the genetic background and the environment. Age, reduced physical activity and obesity represent predominant non-genetic factors. Among genetic factors common variants in a number of type 2 diabetes candidate genes, e.g. PPARG, KCNJ11 and TCF7L2, have been associated with increased risk of the disease. Additionally, more recent whole genome association studies (WGAS) have not only confirmed the previous associated gene regions, but also identified variants in FTO, CDKN2A/CDKN2B, IGF2BP2, CDKAL1, HHEX, WFS1 and SLC30A8 as being associated with type 2 diabetes [9]. We have previously shown that an age-dependent decrease in skeletal muscle PGC-1α and PGC-1β expression is partially heritable and influenced by the PGC-1α Gly482Ser and PGC-1β Ala203Pro polymorphisms [10, 11]. In addition, insulin-resistant offspring of patients with type 2 diabetes were suggested to have impaired mitochondrial function in skeletal muscle [12]. However, the interaction between genes and environment may be even more complex and could involve epigenetic factors such as DNA methylation and histone modifications in the development of type 2 diabetes. Cytosine residues occurring in CG dinucleotides are targets for DNA methylation in vertebrates and gene expression is often reduced when DNA is methylated in the promoter region. We recently showed that such mechanisms can dramatically change expression of another OXPHOS gene, NDUFB6, during a person’s life-time [13].
It is well established that mutations in mtDNA can cause diabetes and other metabolic defects [14–16], but less is known about whether common variations in nuclear encoded OXPHOS genes influence the susceptibility to type 2 diabetes.
Our major aim was to investigate the regulation of OXPHOS gene expression in human skeletal muscle and its role in glucose metabolism and type 2 diabetes. For this purpose we chose the COX7A1 gene, as its expression is decreased in skeletal muscle from patients with type 2 diabetes [3]. Also, the gene resides in a locus on human chromosome 19 showing differential DNA methylation [7]. The specific aims were to: (1) evaluate the impact of genetic (single nucleotide polymorphisms [SNPs]), epigenetic (DNA methylation) and non-genetic factors (e.g. age) on the expression of COX7A1 in human skeletal muscle; and (2) investigate whether common variants in the COX7A1 gene are associated with increased risk of type 2 diabetes.
Methods
Participants
Twins
Participants were identified through The Danish Twin Register. Selection criteria for the young and elderly twins have been previously described and their clinical characteristics are described in Table 1 [10, 17, 18].
The Botnia Study
We studied 1,466 unrelated individuals from the Botnia Study, 751 of whom have been diagnosed with type 2 diabetes at >35 years of age and 715 non-diabetic controls without first-degree relatives with type 2 diabetes (Table 1). The Botnia Study is a family based study established in 1990 aiming at the identification of type 2 diabetes susceptibility genes [19]. Participants were classified into different stages of glucose tolerance based on an OGTT. Individuals with genetically verified MODY or GAD antibody-positive patients were excluded from the study.
Malmö case–control cohort
This included 2,830 Scandinavian type 2 diabetes cases from a local Diabetes Registry [20] and 3,550 unrelated ethnically matched healthy control individuals from the Malmö Diet and Cancer Study [21] (Table 1). Type 2 diabetes was diagnosed according to WHO criteria [22], with C-peptide concentrations ≥0.3 nmol/l, no GAD antibodies and age-at-onset >35 years. The control individuals had fasting blood glucose <5.6 mmol/l and no known family history of type 2 diabetes (Table 1).
All studies were approved by the regional Ethics Committees and conducted according to the Helsinki Declaration.
Clinical examination and muscle biopsies in twins
Twins underwent 2 days of clinical examination including a 2-h euglycaemic–hyperinsulinaemic clamp (40 mU m−2 min−1) combined with indirect calorimetry before (basal) and at the end of the clamp as previously described [10, 17, 18]. Rates of glucose disposal, its partitioning into oxidation and non-oxidative glucose metabolism as well as fat oxidation were calculated as previously reported and expressed as mg (kg lean body mass)−1 min−1 [23]. Muscle biopsies were obtained from the vastus lateralis muscle under local anaesthesia during both basal and insulin-stimulated states. Plasma glucose and insulin concentrations and specific activity of 3H2O were measured as previously described [17, 24, 25].
Analysis of COX7A1 mRNA levels in skeletal muscle
Total RNA was extracted from muscle biopsies using Tri Reagent kit (Sigma-Aldrich, St Louis, MO, USA). cDNA was synthesised using Superscript II RNase H− Reverse Transcriptase and random hexamer primers (Invitrogen, Carlsbad, CA, USA). COX7A1 mRNA levels were quantified by TaqMan real-time PCR with an ABI 7900 system (Applied Biosystems, Foster City, CA, USA) using gene-specific probes and primer pairs for COX7A1 covering exon boundary 1–2 (Assays-on-demands, Hs00156989_m1; Applied Biosystems). Transcript quantity was normalised to the mRNA level of cyclophilin A (4326316E; Applied Biosystems). For each probe/primer set, a standard curve was generated, which was confirmed to increase linearly with increasing amounts of cDNA. The mRNA expression data are expressed as mean ± SEM.
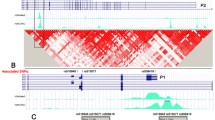
DNA methylation
Genomic DNA was isolated from muscle biopsies at the same time as RNA using the Tri Reagent kit according to the manufacture’s instructions (Sigma-Aldrich). DNA bisulphite modification was accomplished on DNA from ten young and ten elderly twins using the EZ DNA methylation kit (Zymo Research, Orange, CA, USA). Bisulphite-modified DNA was amplified with primers designed using MethPrimer [26] (forward primer 5′-GGGGAGGAGTTGTATTTGTTTT-3′ and reverse primer 5′-CTCTATCCAAAAACCCCAAAC-3′). A second, semi-nested PCR was performed using the forward primer 5′-TTTGTAAAAATGTATTTTTTGGTAT-3′. This amplicon is 238 base pairs and part of the COX7A1 5′ untranslated mRNA region, 64 bases upstream from the first coding exon (Fig. 1b). It includes 22 possible sites subject to DNA methylation (CpG sites) and is situated in a CpG island, i.e. a region of at least 200 base pairs with a GC content greater than 50% and with observed/expected CpG ratio greater than 60% [27]. The PCR products were cloned into plasmid vectors (pCR 4-TOPO; Invitrogen), further Eschericia coli were transformed and DNA from ten colonies of each individual muscle sample were isolated (QIAprep 8 Miniprep Kit; Qiagen, Valencia, CA, USA). The individual clones were sequenced and the number of methylated sites was determined using BiQ Analyser [28]. The proportion of methylation for each individual was calculated by dividing the total number of methylated sites in all clones by the total number of possible methylation sites. Of the 20 samples, six were also directly sequenced following bisulphite treatment of the DNA and analysed using ESME (epigenetic sequencing methylation analysis software; Epigenomics, Berlin, Germany) [29] to verify our results.
Genotyping
Genomic DNA was extracted from blood using conventional methods [30]. Native DNA was used for all genotyping except for the controls in the Malmö cohort, where whole genome amplified DNA (DECODE, Reykjavik, Iceland) was used. Genotype data covering the COX7A1 locus on chr19:41323663-41345611 (10 kb upstream and 10 kb downstream of the COX7A1 gene transcript) (Fig. 1a) was downloaded from the International HapMap project (www.hapmap.org; last accessed March 2008) [31] for the CEPH (Utah residents with ancestry from northern and western Europe) population. Only three SNPs with minor allele frequency >5% were found: rs11665903, rs753420 and rs7255180; the linkage disequilibrium (LD) structure between these SNPs was analysed using Haploview [32]. The LD relationship between the three SNPs was low (r 2 < 0.04) and all the COX7A1 SNPs were therefore selected for genotyping. rs753420 is located in the 5′ mRNA untranslated region, rs11665903 8.4 kb upstream and rs7255180 1.5 kb downstream from the COX7A1 transcript (Fig. 1a). These SNPs were genotyped using a single based extension method adopted for a MassArray (Sequenom) platform in the Profiling Polygenic Disease Swegene laboratory at Lund University, Malmö, Sweden. Genotyping accuracy was determined by 8% re-genotyping with a concordance rate of 99.8% to 100%. To verify the method, a subset of the samples was also analysed using TaqMan SNP Genotyping Assays on the ABI7900 (Applied Biosystems).
Statistical methods
Generalised estimating equations
Because of the strong intrapair correlation of twin data, conventional tests of differences between variable (y) means are not valid. To correct for this dependence, we used generalised estimating equations (GEE) methodology (y = α + βx) to provide valid standard errors for the β-coefficients [33, 34]. The β-coefficient was estimated from all observations, whereas for calculation of the variance of β, each twin pair was considered as one cluster.
Linear regression
To identify factors independently associated with the response variable, we used backward-elimination multivariate regression analysis with p > 0.05 as the defining criterion for exclusion of model terms. Also in this analysis we used GEE methodology to obtain valid tests.
Biometric modelling
The total phenotypic variance is the sum of the variance attributable to the effects of both genetic and environmental factors. The degree of genetic and environmental influence of a variable can be estimated using biometric modelling. The models tested included the following parameters: genetic variance due to additive genetic effects (V A) or dominant genetic effects (V D) and environmental variance due to an individual environment not shared with co-twin (V E) or a common environment shared among co-twins (V C). Biometric modelling was conducted as previously described [35].
Association between genetic variation in the COX7A1 gene and type 2 diabetes
Power to detect association between SNPs in the COX7A1 gene region and type 2 diabetes in our case–control studies was calculated using the Genetic Power Calculator [36]. With a minor allele frequency of 5%, a type 2 diabetes frequency of 6% and a relative risk of 1.3 at α = 0.05, we have 38% power using a dominant model in the Botnia case–control study and a corresponding 93% power in the Malmö case–control study. To determine association of individual SNPs with type 2 diabetes, we performed logistic regression adjusted for BMI, sex and age-at-onset (patients) or age-at-visit (controls), assuming an additive model. Hardy–Weinberg p value, LD and haplotype block structure were calculated using Haploview [32].
DNA methylation
We used a robust rank-order test [37] to test whether the total amount of DNA methylation in the analysed COX7A1 region was increased in elderly compared with young twins. Ten old and ten young twins were analysed for DNA methylation; accordingly results were corrected for inter-twin dependence. Power to detect differences in DNA methylation was calculated to 73.9% (α = 0.05) using DSS research statistical power calculator (www.dssresearch.com; Fort Worth, TX, USA).
Results
Heritability of COX7A1 mRNA expression in skeletal muscle of twins
The characteristics of the twins are described in Table 1. The degree of genetic and environmental influence on muscle COX7A1 mRNA expression in skeletal muscle was estimated by biometric modelling in twins. The heritability was estimated to be 50% in young and 72% in elderly twins, suggesting that both genetic and environmental factors influence COX7A1 expression in muscle (Electronic supplementary material [ESM] Table 1).
COX7A1 mRNA expression in skeletal muscle declines with age
The mRNA level of COX7A1 was decreased in skeletal muscle from elderly (n = 68) compared with young (n = 88) twins, both in the basal and insulin-stimulated state (basal 1.00 ± 0.05 vs 1.68 ± 0.06 and clamp 1.04 ± 0.05 vs 1.71 ± 0.06; p = 0.0005) (Fig. 2). The euglycaemic–hyperinsulinaemic clamp did not influence the mRNA level of COX7A1 either in young or in elderly twins (Fig. 2).
Effects of age and insulin on human skeletal muscle COX7A1 mRNA levels in 88 young and 68 elderly twins, before (white bars) and after (black bars) a euglycaemic–hyperinsulinaemic clamp. RNA was analysed for COX7A1 mRNA expression together with the endogenous control cyclophilin A. The COX7A1:cyclophilin A ratio was calculated for each sample and the ratios are presented. Results are expressed as the mean ± SEM. *p ≤ 0.05
The level of DNA methylation in the COX7A1 promoter region increases with age
DNA methylation in the COX7A1 promoter region was analysed in genomic DNA extracted from skeletal muscle of ten elderly and ten young twins. Figure 1b shows the location of the analysed sequence in the proposed COX7A1 promoter, a sequence within a CpG island including 22 methylation sites. The degree of DNA methylation was higher in elderly than in young twins (19.9 ± 8.3% vs 1.8 ± 2.7%, p = 0.035; data median ± SEM). Furthermore, a box-plot illustrating the variation in DNA methylation between individuals suggests that differences in DNA methylation become more pronounced with increasing age (Fig. 3). However, we did not observe any significant correlation between COX7A1 DNA methylation and mRNA expression (r = −0.33, p = 0.15).
Genetic variation influences COX7A1 expression in muscle
The heritability estimates demonstrated that both genetic and non-genetic factors influence the expression level of COX7A1 in muscle (ESM Table 1). In order to evaluate the genetic factors influencing COX7A1 expression, three SNPs were genotyped in the twins and related to COX7A1 mRNA expression in skeletal muscle. One polymorphism, rs753420, influenced basal COX7A1 expression in muscle of young (T/T [n = 39] 1.53 ± 0.08, T/G [n = 42] 1.71 ± 0.08 vs G/G [n = 7] 2.3 ± 0.19; p = 0.0001) but not of elderly twins (Table 2). SNPs rs11665903 and rs7255180 did not significantly influence basal COX7A1 expression in either of the young or elderly twin groups (Table 2).
COX7A1 polymorphisms and risk of type 2 diabetes
We also investigated whether common variants in the COX7A1 gene are associated with increased risk of type 2 diabetes in two independent case–control cohorts (Table 1). None of the three SNPs showed significant association to type 2 diabetes in the Botnia cohort. In the Malmö case–control cohort, one SNP (rs7255180) showed a nominal association to type 2 diabetes (OR 0.80 [0.66–0.97], p = 0.027) for an additive model, but this did not stand correction for multiple testing (Table 3). A heterogeneity test excluded significant difference between the Botnia and the Malmö cohorts (p = 0.77), and to increase power, we pooled our data from the two cohorts. However, even after pooling the two cohorts, we could not detect any genotype association to type 2 diabetes (Table 3).
PGC-1α is associated with COX7A1 mRNA expression in human skeletal muscle
A multivariate regression analysis was performed to test whether any of the following factors could influence COX7A1 mRNA levels in skeletal muscle of the twins: PGC-1α mRNA expression in skeletal muscle, age, sex and BMI (ESM Table 2). The final model was reached using backward selection regression. The level of COX7A1 was positively associated with PGC-1α expression (regression coefficient = 0.27; p = 0.003) and negatively associated with increased age (regression coefficient = −0.52; p < 0.0001) and female sex (regression coefficient = −0.21; p = 0.01; ESM Table 2).
COX7A1 expression is associated with in vivo glucose uptake and total body aerobic capacity in twins
We also tested whether COX7A1 mRNA expression in skeletal muscle was associated with insulin-stimulated glucose uptake and total body aerobic capacity (\(\mathop V\limits^ \cdot {\text{O}}_{{\text{2max}}} \)) using multivariate regression analysis including age, sex and BMI as covariates (Table 4). The level of COX7A1 was positively related with glucose uptake (p = 0.01) and \(\mathop V\limits^ \cdot {\text{O}}_{{\text{2max}}} \) (p = 0.001).
Discussion
The key findings of the present study were that age influences both promoter DNA methylation and gene expression of COX7A1 in human skeletal muscle (Fig. 4). Our results demonstrate how the genome can be modified during life by DNA methylation, which theoretically could influence gene expression and thereby mitochondrial metabolism. It was also shown that the transcript level of this OXPHOS gene was positively associated with in vivo glucose uptake and \(\mathop V\limits^ \cdot {\text{O}}_{{\text{2max}}} \), although we were unable to detect any significant association between COX7A1 polymorphisms and type 2 diabetes.
Proposed mechanisms for regulation of COX7A1 in human skeletal muscle and its effect on in vivo metabolism. According to our findings, age increases DNA methylation and decreases COX7A1 mRNA expression. Furthermore, COX7A1 mRNA expression is associated with genetic variation (SNP rs753420) and PGC-1α mRNA expression. As a result this could affect in vivo metabolism, as COX7A1 mRNA expression was positively associated with both insulin-stimulated glucose uptake and \(\mathop V\limits^ \cdot {\text{O}}_{{\text{2max}}} \)
COX7A1 was chosen as a candidate because it is among the PGC-1α-responsive genes that are involved in OXPHOS and are coordinately downregulated in human diabetes [3]. A close relationship between COX7A1 and PGC-1α expression was confirmed in the present study.
The prevalence of type 2 diabetes increases with age and is increased in individuals with insulin resistance. Oxidative capacity in skeletal muscle has been shown to be a good predictor of insulin sensitivity [38]. We analysed the expression level of COX7A1 in muscle biopsies from young and elderly non-diabetic twins to determine whether the level of this OXPHOS gene declines with age and is related to insulin sensitivity in vivo. The COX7A1 mRNA level was reduced by 40% in the elderly twins, as compared with the 23% decrease observed in muscle of patients with type 2 diabetes [3]. A decline in COX7A1 with age may contribute to increased insulin resistance, since elderly twins were more insulin-resistant than younger twins and COX7A1 expression in muscle was associated with insulin-stimulated glucose uptake. Our results are thus in line with other reports demonstrating a decrease in mitochondrial OXPHOS activity [39] or expression of OXPHOS genes and insulin-stimulated glucose uptake in muscle of elderly individuals [13, 40, 41]. Collectively these studies demonstrate that age has a strong influence on expression of OXPHOS genes in human skeletal muscle.
Expression of COX7A1 showed clear heritability in both young and elderly twins suggesting that OXPHOS expression is under genetic control. In support of this, the SNP rs753420 was associated with basal COX7A1 mRNA expression in muscle. However, this did not translate into an increased risk of type 2 diabetes, as there was no significant association between SNPs in and around the COX7A1 gene and type 2 diabetes. Although the case–control study was fairly well powered for the Malmö cohort, we cannot exclude association with lower odds ratios than 1.3. Recently a meta-analysis based on three type 2 diabetes WGAS was performed [42]. The results include data from 10,128 individuals and ~2.2 million SNPs. None of the three COX7A1 polymorphisms investigated in our study showed association with type 2 diabetes in this meta-analysis.
It is assumed that DNA methylation of specific sites, particularly in the promoter region, may suppress the expression of the gene [43, 44]. Although the expression of COX7A1 is tissue-specific, this gene is located in a CpG island, a feature which is commonly seen among housekeeping genes [6]. CpG islands are regions characterised by high C + G and CpG content, found in about 60% of human gene promoters. They are mainly unmethylated, although a subset can show tissue-specific methylation during development [45]. We recently showed increased DNA methylation in another OXPHOS gene, NDUFB6, in muscle of elderly compared with that of young twins; this increase in DNA methylation was associated with an age-related decrease in gene expression [13]. We therefore investigated whether this was also true for the COX7A1 gene region.
Genomic DNA isolated from skeletal muscle was used to analyse DNA methylation in a region of the COX7A1 gene previously suggested to be important for initiation of transcription [6, 46]. We detected an increased mean methylation rate in elderly compared with young twins. Also, there was more variation within the group of the elderly twins, suggesting that the variation in DNA methylation between individuals becomes more pronounced with increasing age. Our findings are supported by previous results showing accumulation of epigenetic alterations with ageing [47–49]. Chalaya et al. [7] have previously compared the DNA methylation patterns in selected promoter regions in the FXYD5–COX7A1 locus in relation to gene expression in human cell lines and tissues (cerebellum, liver, kidney and stomach). Whereas the housekeeping gene COX6B (also known as COX6B1) showed a clear correlation between the level of DNA methylation and transcription level, others did not.
We could not detect any CpG sites more prone to methylation than others; therefore we calculated the mean methylation level for each individual rather than analysing each specific methylation site. Unlike in our previous study [13], we did not observe a significant correlation (r = −0.33) between the level of DNA methylation and gene expression of COX7A1. This could be due to low power or the large variation in DNA methylation observed in the elderly, suggesting that non-genetic factors also might influence the age-related decline in COX7A1 expression. However, the present study, together with our previous data [13, 50], does show that age can alter the level of DNA methylation, which theoretically then could influence gene expression and thereby metabolism. Furthermore, no known polymorphisms in the COX7A1 gene region affect methylation sites.
In conclusion, we observed an age-related increase in DNA methylation of COX7A1 as well as a decrease in expression of this gene in skeletal muscle of elderly individuals. Our data also propose a role for COX7A1 in regulating in vivo glucose uptake and \(\mathop V\limits^ \cdot {\text{O}}_{{\text{2max}}} \) (Fig. 4).
Abbreviations
- COX:
-
cytochrome c oxidase
- GEE:
-
generalised estimating equations
- LD:
-
linkage disequilibrium
- mtDNA:
-
mitochondrial DNA
- OXPHOS:
-
oxidative phosphorylation
- SNP:
-
single nucleotide polymorphism
- \(\mathop V\limits^ \cdot {\text{O}}_{{\text{2max}}} \) :
-
total body aerobic capacity
- WGAS:
-
whole genome association study
References
Kelley DE, He J, Menshikova EV, Ritov VB (2002) Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 51:2944–2950
Huang X, Eriksson KF, Vaag A et al (1999) Insulin-regulated mitochondrial gene expression is associated with glucose flux in human skeletal muscle. Diabetes 48:1508–1514
Mootha VK, Lindgren CM, Eriksson KF et al (2003) PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34:267–273
Patti ME, Butte AJ, Crunkhorn S et al (2003) Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci USA 100:8466–8471
Sreekumar R, Halvatsiotis P, Schimke JC, Nair KS (2002) Gene expression profile in skeletal muscle of type 2 diabetes and the effect of insulin treatment. Diabetes 51:1913–1920
Wolz W, Kress W, Mueller CR (1997) Genomic sequence and organization of the human gene for cytochrome c oxidase subunit (COX7A1) VIIa-M. Genomics 45:438–442
Chalaya TV, Akopov SB, Nikolaev LG, Sverdlov ED (2006) Tissue specificity of methylation of cytosines in regulatory regions of four genes located in the locus FXYD5-COX7A1 of human chromosome 19: correlation with their expression level. Biochemistry 71:294–299
Arnaudo E, Hirano M, Seelan RS et al (1992) Tissue-specific expression and chromosome assignment of genes specifying two isoforms of subunit VIIa of human cytochrome c oxidase. Gene 119:299–305
Frayling TM (2007) Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat Rev 8:657–662
Ling C, Poulsen P, Carlsson E et al (2004) Multiple environmental and genetic factors influence skeletal muscle PGC-1α and PGC-1β gene expression in twins. J Clin Invest 114:1518–1526
Ling C, Wegner L, Andersen G et al (2007) Impact of the peroxisome proliferator activated receptor-gamma coactivator-1β (PGC-1β) Ala203Pro polymorphism on in vivo metabolism, PGC-1β expression and fibre type composition in human skeletal muscle. Diabetologia 50:1615–1620
Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI (2004) Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med 350:664–671
Ling C, Poulsen P, Simonsson S et al (2007) Genetic and epigenetic factors are associated with expression of respiratory chain component NDUFB6 in human skeletal muscle. J Clin Invest 117:3427–3435
Poulton J, Luan J, Macaulay V, Hennings S, Mitchell J, Wareham NJ (2002) Type 2 diabetes is associated with a common mitochondrial variant: evidence from a population-based case-control study. Hum Mol Genet 11:1581–1583
van den Ouweland JM, Lemkes HH, Ruitenbeek W et al (1992) Mutation in mitochondrial tRNA(Leu)(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nat Genet 1:368–371
Wilson FH, Hariri A, Farhi A et al (2004) A cluster of metabolic defects caused by mutation in a mitochondrial tRNA. Science 306:1190–1194
Poulsen P, Levin K, Beck-Nielsen H, Vaag A (2002) Age-dependent impact of zygosity and birth weight on insulin secretion and insulin action in twins. Diabetologia 45:1649–1657
Poulsen P, Vaag A (2006) The intrauterine environment as reflected by birth size and twin and zygosity status influences insulin action and intracellular glucose metabolism in an age- or time-dependent manner. Diabetes 55:1819–1825
Groop L, Forsblom C, Lehtovirta M et al (1996) Metabolic consequences of a family history of NIDDM (the Botnia Study): evidence for sex-specific parental effects. Diabetes 45:1585–1593
Lindholm E, Agardh E, Tuomi T, Groop L, Agardh CD (2001) Classifying diabetes according to the new WHO clinical stages. Eur J Epidemiol 17:983–989
Leosdottir M, Nilsson P, Nilsson JA, Mansson H, Berglund G (2004) The association between total energy intake and early mortality: data from the Malmo Diet and Cancer Study. J Intern Med 256:499–509
Alberti KG, Zimmet PZ (1998) Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med 15:539–553
Poulsen P, Levin K, Petersen I, Christensen K, Beck-Nielsen H, Vaag A (2005) Heritability of insulin secretion, peripheral and hepatic insulin action, and intracellular glucose partitioning in young and old Danish twins. Diabetes 54:275–283
Hemmila I, Dakubu S, Mukkala VM, Siitari H, Lovgren T (1984) Europium as a label in time-resolved immunofluorometric assays. Anal Biochem 137:335–343
Hother-Nielsen O, Beck-Nielsen H (1990) On the determination of basal glucose production rate in patients with type 2 (non-insulin-dependent) diabetes mellitus using primed-continuous 3-3H-glucose infusion. Diabetologia 33:603–610
Li LC, Dahiya R (2002) MethPrimer: designing primers for methylation PCRs. Bioinformatics (Oxford, England) 18:1427–1431
Gardiner-Garden M, Frommer M (1987) CpG islands in vertebrate genomes. J Mol Biol 196:261–282
Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T (2005) BiQ analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics (Oxford, England) 21:4067–4068
Lewin J, Schmitt AO, Adorjan P, Hildmann T, Piepenbrock C (2004) Quantitative DNA methylation analysis based on four-dye trace data from direct sequencing of PCR amplificates. Bioinformatics 20:3005–3012
Vandenplas S, Wiid I, Grobler-Rabie A et al (1984) Blot hybridisation analysis of genomic DNA. J Med Genet 21:164–172
The International HapMap Consortium (2003) The International HapMap Project. Nature 426:789–796
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics (Oxford, England) 21:263–265
Liang KY, Zeger SL (1993) Regression analysis for correlated data. Annu Rev Public Health 14:43–68
Zeger SL, Liang KY (1986) Longitudinal data analysis for discrete and continuous outcomes. Biometrics 42:121–130
Storgaard H, Poulsen P, Ling C, Groop L, Vaag AA (2006) Genetic and nongenetic determinants of skeletal muscle glucose transporter 4 messenger ribonucleic acid levels and insulin action in twins. J Clin Endocrinol Metab 91:702–708
Purcell S, Cherny SS, Sham PC (2003) Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19:149–150
Siegel S, Castellan NJJ (1988) Nonparametric statistics for the behavioral sciences. McGraw-Hill International, Columbus, OH
Bruce CR, Anderson MJ, Carey AL et al (2003) Muscle oxidative capacity is a better predictor of insulin sensitivity than lipid status. J Clin Endocrinol Metab 88:5444–5451
Petersen KF, Befroy D, Dufour S et al (2003) Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science 300:1140–1142
Trounce I, Byrne E, Marzuki S (1989) Decline in skeletal muscle mitochondrial respiratory chain function: possible factor in ageing. Lancet 1:637–639
Zahn JM, Sonu R, Vogel H et al (2006) Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Genet 2:e115
Zeggini E, Scott LJ, Saxena R, Voight BF for the DIAGRAM consortium (2008) Meta-analysis of genome-wide association data and large-scale replication identifies several additional susceptibility loci for type 2 diabetes. Nat Genet DOI 10.1038/ng.120
Eckhardt F, Lewin J, Cortese R et al (2006) DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet 38:1378–1385
Weber M, Hellmann I, Stadler MB et al (2007) Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39:457–466
Bernstein BE, Meissner A, Lander ES (2007) The mammalian epigenome. Cell 128:669–681
Yu M, Jaradat SA, Grossman LI (2002) Genomic organization and promoter regulation of human cytochrome c oxidase subunit VII heart/muscle isoform (COX7AH). Biochim Biophys Acta 1574:345–353
Fraga MF, Esteller M (2007) Epigenetics and aging: the targets and the marks. Trends Genet 23:413–418
Poulsen P, Esteller M, Vaag A, Fraga MF (2007) The epigenetic basis of twin discordance in age-related diseases. Pediatr Res 61:38R–42R
Richardson BC (2002) Role of DNA methylation in the regulation of cell function: autoimmunity, aging and cancer. J Nutr 132:2401S–2405S
Fraga MF, Ballestar E, Paz MF et al (2005) Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA 102:10604–10609
Acknowledgements
This study was supported by grants from the Danish Medical Research Council, Danish Diabetes Association, Region Skåne, Malmö University Hospital, Swegene, the Diabetes Programme at Lund University, Novo Nordisk, Crafoord, Hedlund, Bergvall, Wiberg, Påhlsson, Hains, Lundberg, Exgenesis and a Linne grant (B31 5631/2006), as well as a grant 522-2006-6480 from the Swedish Research Council. We thank E. Nilsson, M. Modest and M. Svensson for technical assistance. We also thank E. Zeggini, L. J. Scott, R. Saxena and B. F. Voight of the Diabetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium for sharing data.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material
ESM Table 1
Best fitting biometric models for basal mRNA COX7A1 expression in muscle from 88 young and 68 elderly twins (PDF 74 KB)
ESM Table 2
Non-genetic factors correlating with basal COX7A1 mRNA expression in skeletal muscle of 88 young and 68 elderly twins (PDF 70 KB)
Rights and permissions
About this article
Cite this article
Rönn, T., Poulsen, P., Hansson, O. et al. Age influences DNA methylation and gene expression of COX7A1 in human skeletal muscle. Diabetologia 51, 1159–1168 (2008). https://doi.org/10.1007/s00125-008-1018-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-008-1018-8