Abstract
Key message
Trait-assisted genomic prediction approach is a way to improve genetic gain by cost unit, by reducing budget allocated to phenotyping or by increasing the program’s size for the same budget.
Abstract
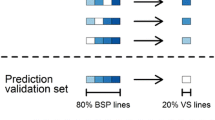
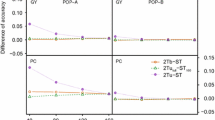
This study compares different strategies of genomic prediction to optimize resource allocation in breeding schemes by using information from cheaper correlated traits to predict a more expensive trait of interest. We used bread wheat baking score (BMS) calculated for French registration as a case study. To conduct this project, 398 lines from a public breeding program were genotyped and phenotyped for BMS and correlated traits in 11 locations in France between 2000 and 2016. Single-trait (ST), multi-trait (MT) and trait-assisted (TA) strategies were compared in terms of predictive ability and cost. In MT and TA strategies, information from dough strength (W), a cheaper trait correlated with BMS (r = 0.45), was evaluated in the training population or in both the training and the validation sets, respectively. TA models allowed to reduce the budget allocated to phenotyping by up to 65% while maintaining the predictive ability of BMS. TA models also improved the predictive ability of BMS compared to ST models for a fixed budget (maximum gain: + 0.14 in cross-validation and + 0.21 in forward prediction). We also demonstrated that the budget can be further reduced by approximately one fourth while maintaining the same predictive ability by reducing the number of phenotypic records to estimate BMS adjusted means. In addition, we showed that the choice of the lines to be phenotyped can be optimized to minimize cost or maximize predictive ability. To do so, we extended the mean of the generalized coefficient of determination (CDmean) criterion to the multi-trait context (CDmulti).





Similar content being viewed by others
References
Akdemir D, Sanchez JI, Jannink J-L (2015) Optimization of genomic selection training populations with a genetic algorithm. GenetSelEvol 47:38. https://doi.org/10.1186/s12711-015-0116-6
Anjum FM, Khan MR, Din A et al (2007) Wheat Gluten: high molecular weight glutenin subunits—structure, genetics, and relation to dough elasticity. J Food Sci 72:R56–R63. https://doi.org/10.1111/j.1750-3841.2007.00292.x
Arruda MP, Lipka AE, Brown PJ et al (2016) Comparing genomic selection and marker-assisted selection for Fusarium head blight resistance in wheat (Triticum aestivum L.). Mol Breed 36:84. https://doi.org/10.1007/s11032-016-0508-5
Bao Y, Kurle JE, Anderson G et al (2015) Association mapping and genomic prediction for resistance to sudden death syndrome in early maturing soybean germplasm. Mol Breed 35(6):128
Beavis WD (1998) QTL analyses: power, precision, and accuracy. In: Paterson AH (ed) Molecular dissection of complex traits. CRC Press, New York, pp 145–162
Bernardo R (2014) Genomewide selection when major genes are known. Crop Sci 54:68–75. https://doi.org/10.2135/cropsci2013.05.0315
Butler DG, Cullis BR, Gilmour AR, Gogel BJ (2009) ASReml-r reference manual. The State of Queensland, Department of Primary Industries and Fisheries, Brisbane
Calus MP, Veerkamp RF (2011) Accuracy of multi-trait genomic selection using different methods. Genet SelEvol 43:26. https://doi.org/10.1186/1297-9686-43-26
Crain J, Mondal S, Rutkoski J et al (2018) Combining high-throughput phenotyping and genomic information to increase prediction and selection accuracy in wheat breeding. Plant Genome. https://doi.org/10.3835/plantgenome2017.05.0043
Crossa J, Pérez P, Hickey J et al (2014) Genomic prediction in CIMMYT maize and wheat breeding programs. Heredity 112:48–60. https://doi.org/10.1038/hdy.2013.16
dos Santos JPR, de Vasconcellos RC, C, Pires LPM, et al (2016) inclusion of dominance effects in the multivariate GBLUP model. PLoS ONE 11:e0152045. https://doi.org/10.1371/journal.pone.0152045
Endelman JB (2011) Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 4:250–255. https://doi.org/10.3835/plantgenome2011.08.0024
Endelman JB, Jannink JL (2012) Shrinkage estimation of the realized relationship matrix. G3 2:1405–1413. https://doi.org/10.1534/g3.112.004259
Fernandes SB, Dias KOG, Ferreira DF, Brown PJ (2018) Efficiency of multi-trait, indirect, and trait-assisted genomic selection for improvement of biomass sorghum. Theor Appl Genet 131:747–755. https://doi.org/10.1007/s00122-017-3033-y
Fisher RA (1919) XV.—The Correlation between Relatives on the Supposition of Mendelian Inheritance. Earth Environ Sci Trans R Soc Edinb 52:399–433. https://doi.org/10.1017/S0080456800012163
Guo G, Zhao F, Wang Y et al (2014) Comparison of single-trait and multiple-trait genomic prediction models. BMC Genet 15:30. https://doi.org/10.1186/1471-2156-15-30
Habier D, Fernando RL, Dekkers JCM (2007) The impact of genetic relationship information on genome-assisted breeding values. Genetics 177:2389–2397. https://doi.org/10.1534/genetics.107.081190
Hayashi T, Iwata H (2013) A Bayesian method and its variational approximation for prediction of genomic breeding values in multiple traits. BMC Bioinform 14:34. https://doi.org/10.1186/1471-2105-14-34
Hayes BJ, Panozzo J, Walker CK et al (2017) Accelerating wheat breeding for end-use quality with multi-trait genomic predictions incorporating near infrared and nuclear magnetic resonance-derived phenotypes. Theor Appl Genet 130:2505–2519. https://doi.org/10.1007/s00122-017-2972-7
Henderson CR, Quaas RL (1976) Multiple trait evaluation using relatives’ records. J Anim Sci 43:1188–1197. https://doi.org/10.2527/jas1976.4361188x
Heslot N, Feoktistov V (2017) Optimization of selective phenotyping and population design for genomic prediction. BioRxiv, 172064.
Isidro J, Jannink J-L, Akdemir D et al (2015) Training set optimization under population structure in genomic selection. Theor Appl Genet 128:145–158. https://doi.org/10.1007/s00122-014-2418-4
Jia Y, Jannink J-L (2012) Multiple-trait genomic selection methods increase genetic value prediction accuracy. Genetics 192:1513–1522. https://doi.org/10.1534/genetics.112.144246
Lado B, Vázquez D, Quincke M et al (2018) Resource allocation optimization with multi-trait genomic prediction for bread wheat (Triticum aestivum L.) baking quality. Theor Appl Genet 131:2719–2731. https://doi.org/10.1007/s00122-018-3186-3
Laloë D (1993) Precision and information in linear models of genetic evaluation. Genet SelEvol 25(6):557
Lande R, Thompson R (1990) Efficiency of marker-assisted selection in the improvement of quantitative traits. Genetics 124:743–756
Liu Y, He Z, Appels R, Xia X (2012) Functional markers in wheat: current status and future prospects. Theor Appl Genet 125:1–10. https://doi.org/10.1007/s00122-012-1829-3
Lynch M, Walsh B (1998) Genetics and analysis of quantitative traits. Sinauer, Sunderland, MA
Meuwissen TH, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Michel S, Kummer C, Gallee M et al (2018) Improving the baking quality of bread wheat by genomic selection in early generations. Theor Appl Genet 131:477–493. https://doi.org/10.1007/s00122-017-2998-x
Montesinos-López OA, Montesinos-López A, Crossa J et al (2016) A genomic Bayesian multi-trait and multi-environment model. G3 6(9):2725–2744
Oury F-X, Chiron H, Faye A et al (2009) The prediction of bread wheat quality: joint use of the phenotypic information brought by technological tests and the genetic information brought by HMW and LMW glutenin subunits. Euphytica 171:87. https://doi.org/10.1007/s10681-009-9997-1
Payne PI (1987) Genetics of wheat storage proteins and the effect of allelic variation on bread-making quality. Ann Rev Plant Physiol 38:141–153. https://doi.org/10.1146/annurev.pp.38.060187.001041
Poland J, Endelman J et al (2012) Genomic selection in wheat breeding using genotyping-by-sequencing. plant. Genome 5:103–113. https://doi.org/10.3835/plantgenome2012.06.0006
Ravel C, Faye A, Ben-Sadoun S et al (2020) SNP markers for early identification of high molecular weight glutenin subunits (HMW-GSs) in bread wheat. TheorAppl Genet. https://doi.org/10.1007/s00122-019-03505-y
Rimbert H, Darrier B, Navarro J et al (2018) High throughput SNP discovery and genotyping in hexaploid wheat. PLoS ONE 13:e0186329. https://doi.org/10.1371/journal.pone.0186329
Rincent R, Charcosset A, Moreau L (2017) Predicting genomic selection efficiency to optimize calibration set and to assess prediction accuracy in highly structured populations. Theor Appl Genet 130:2231–2247. https://doi.org/10.1007/s00122-017-2956-7
Rincent R, Laloë D, Nicolas S et al (2012) Maximizing the reliability of genomic selection by optimizing the calibration set of reference individuals: comparison of methods in two diverse groups of maize inbreds (Zea mays L.). Genetics 192:715–728. https://doi.org/10.1534/genetics.112.141473
Rutkoski J, Poland J, Mondal S et al (2016) canopy temperature and vegetation indices from high-throughput phenotyping improve accuracy of pedigree and genomic selection for grain yield in wheat. G3 6:2799–2808. https://doi.org/10.1534/g3.116.032888
Sarinelli JM, Murphy JP, Tyagi P et al (2019) Training population selection and use of fixed effects to optimize genomic predictions in a historical USA winter wheat panel. Theor Appl Genet 132:1247–1261. https://doi.org/10.1007/s00122-019-03276-6
Schulthess AW, Wang Y, Miedaner T et al (2016) Multiple-trait- and selection indices-genomic predictions for grain yield and protein content in rye for feeding purposes. Theor Appl Genet 129:273–287. https://doi.org/10.1007/s00122-015-2626-6
Schulthess AW, Zhao Y, Longin CFH, Reif JC (2018) Advantages and limitations of multiple-trait genomic prediction for Fusarium head blight severity in hybrid wheat (Triticum aestivum L.). Theor Appl Genet 131:685–701. https://doi.org/10.1007/s00122-017-3029-7
Shewry PR (2009) Wheat. J Exp Bot 60:1537–1553. https://doi.org/10.1093/jxb/erp058
Storlie E, Charmet G (2013) Genomic selection accuracy using historical data generated in a wheat breeding program. Plant Genome. https://doi.org/10.3835/plantgenome2013.01.0001
Sun J, Rutkoski JE, Poland JA et al (2017) Multitrait, random regression, or simple repeatability model in high-throughput phenotyping data improve genomic prediction for wheat grain yield. Plant Genome. https://doi.org/10.3835/plantgenome2016.11.0111
Venables WN, Ripley BD (2002) Modern applied statistics with S. fourth edition. Springer, New York. ISBN 0-387-95457-0
Whittaker JC, Thompson R, Denham MC (2000) Marker-assisted selection using ridge regression. Genet Res 75:249–252. https://doi.org/10.1017/S0016672399004462
Acknowledgements
The authors thank the genotyping platform GENTYANE at INRA Clermont-Ferrand (gentyane.clermont.inra.fr) which has conducted the genotyping. The work in experimental units by INRA (Clermont-Ferrand, Dijon, Estrées-Mons, Lusignan, Le Moulon and Rennes) and in Agri-Obtentions is also gratefully acknowledged. Doctoral work of SBS was funded by a grant from the Auvergne-Rhônes-Alpes region and from the European Regional Development Fund (FEDER). This work was supported by the Breed wheat project thanks to the funding from the French Government managed by the National Research Agency (ANR) in the framework of Investments for the Future (ANR-10-BTBR-03) France AgriMer and the French Fund to support Plant Breeding (FSOV).
Author information
Authors and Affiliations
Contributions
JA, FXO, BR and EH designed the field trials and collected the phenotypic data. RR supported in statistical analysis and in developing the multi-trait CDmean algorithm. CR developed KASP markers derived from HMW-GS loci. SBS analyzed the data and wrote the manuscript. SB and GG guided through the study and helped improving the manuscript. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by Hiroyoshi Iwata.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendix
Appendix
We extended here the generalized CD (Laloë 1993) to the multi-trait context. The objective is to compute the expected reliability (before phenotyping) in a multi-trait context for contrasts corresponding to the prediction objectives. To give an example, it is clear that if the objective is to accurately predict the difference (contrasts) between families, or to focus on predictions within families, the contrasts to be considered will be different and the optimal calibration sets as well. By defining contrasts corresponding to the prediction objectives, one can adapt the CDmulti criterion to the specific prediction objectives (see Rincent et al. 2017 for more details in the single-trait context).
For a given contrast c, the generalized prediction error variance (PEV(c)) in the multi-trait context is equal to:
where X and Z are the design matrices for the fixed and random effects, respectively, K is the kinship matrix, Σa is the genetic variance–covariance matrix between traits, and Σε is the residual variance–covariance matrix between traits. ⊗ indicate the Kronecker product operator between matrices.
The generalized multi-trait CD for a given contrast c is equal to:
As a reminder, a contrast is a vector whose elements sum to 0 and indicating the difference in which we are interested. For example, if we are interested in accurately predicting the difference between individual 1 and individual 2, the contrast to consider will be: \(c^{T} = \left[ {1, - 1,0,0,0,0, \ldots } \right]\).
Rights and permissions
About this article
Cite this article
Ben-Sadoun, S., Rincent, R., Auzanneau, J. et al. Economical optimization of a breeding scheme by selective phenotyping of the calibration set in a multi-trait context: application to bread making quality. Theor Appl Genet 133, 2197–2212 (2020). https://doi.org/10.1007/s00122-020-03590-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-020-03590-4