Abstract
Key message
A wild rice QTL qGL12.2 for grain length was fine mapped to an 82-kb interval in chromosome 12 containing six candidate genes and none was reported previously.
Abstract
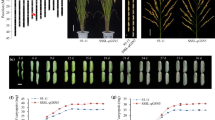
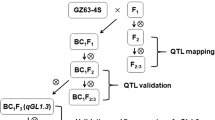
Grain length is an important trait for yield and commercial value in rice. Wild rice seeds have a very slender shape and have many desirable genes that have been lost in cultivated rice during domestication. In this study, we identified a quantitative trait locus, qGL12.2, which controls grain length in wild rice. First, a wild rice chromosome segment substitution line, CSSL41, was selected that has longer glume and grains than does the Oryza sativa indica cultivar, 9311. Next, an F2 population was constructed from a cross between CSSL41 and 9311. Using the next-generation sequencing combined with bulked-segregant analysis and F3 recombinants analysis, qGL12.2 was finally fine mapped to an 82-kb interval in chromosome 12. Six candidate genes were found, and no reported grain length genes were found in this interval. Using scanning electron microscopy, we found that CSSL41 cells are significantly longer than those of 9311, but there is no difference in cell widths. These data suggest that qGL12.2 is a novel gene that controls grain cell length in wild rice. Our study provides a new genetic resource for rice breeding and a starting point for functional characterization of the wild rice GL gene.






Similar content being viewed by others
References
Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H et al (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30:174178
Ali ML, Sanchez PL, Sb Yu, Lorieux M, Eizenga GC (2010) Chromosome segment substitution lines: a powerful tool for the introgression of valuable genes from Oryza wild species into cultivated rice (O. sativa). Rice 3:218–234
Chen J, Wang ZY (2002) Progress in the study of plant MYB transcription factors. J Plant Physiol Mol Biol 28:81–88
Fan C, Xing Y, Mao H, Lu T, Han B, Xu C et al (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet 112:1164–1671
Frary A, Nesbitt TC, Aea Frary (2000) fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289:85–88
Guo G, Wang S, Liu J, Pan B, Diao W, Ge W et al (2017) Rapid identification of QTLs underlying resistance to Cucumber mosaic virus in pepper (Capsicum frutescens). Theor Appl Genet 130:41–52
Han Y, Lv P, Hou S, Li S, Ji G, Ma X et al (2015) Combining next generation sequencing with bulked segregant analysis to fine map a stem moisture locus in Sorghum (Sorghum bicolor L. Moench). PLoS One 10:e0127065
Hartwig B, James GV, Konrad K, Schneeberger K, Turck F (2012) Fast isogenic mapping-by-sequencing of ethyl methanesulfonate-induced mutant bulks. Plant Physiol 160:591–600
Heang D, Sassa H (2012) Antagonistic actions of HLH/bHLH proteins are involved in grain length and weight in rice. PLoS One 7:e31325
Hu J, Wang Y, Fang Y, Zeng L, Xu J, Yu H et al (2015) A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant 8:1455–1465
Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y et al (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42:961–967
Huang X, Kurata N, Wei X, Wang ZX, Wang A, Zhao Q et al (2012) A map of rice genome variation reveals the origin of cultivated rice. Nature 490:497–501
Huang R, Jiang L, Zheng J, Wang T, Wang H, Huang Y et al (2013) Genetic bases of rice grain shape: so many genes, so little known. Trends Plant Sci 18:218–226
International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800
Juliano BO, Villareal CP (1993) Grain quality evaluation of world rices. International Rice Research Institute, Manila
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760
Li Y, Fan C, Xing Y, Jiang Y, Luo L, Sun L et al (2011) Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet 43:1266–1269
Lin LH, Wu WR (2003) Mapping of QTLs underlying grain shape and grain weight in rice. Mol Plant Breed 1:337–342
Lu H, Lin T, Klein J, Wang S, Qi J, Zhou Q et al (2014) QTL-seq identifies an early flowering QTL located near flowering locus T in cucumber. Theor Appl Genet 127:1491–1499
McCouch SR, Sweeney M, Li J, Jiang H, Thomson M, Septiningsih E et al (2007) Through the genetic bottleneck: O. rufipogon as a source of trait-enhancing alleles for O. sativa. Euphytica 154:317–339
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A et al (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
Meng L, Li H, Zhang L, Wang J (2015) QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 3:269–283
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci 88:9828–9832
Panaud O, Chen X, McCouch SR (1996) Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.). Mol Gen Genet 252:597–607
Qi L, Sun Y, Li J, Su L, Zheng XM, Wang XN et al (2017) Identify QTLs for grain size and weight in common wild rice using chromosome segment substitution lines across six environments. Breed Sci. https://doi.org/10.1270/jsbbs.16082
Qiao W, Qi L, Cheng Z, Su L, Li J, Sun Y et al (2016) Development and characterization of chromosome segment substitution lines derived from Oryza rufipogon in the genetic background of O. sativa spp. indica cultivar 9311. BMC Genom 17:580
Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL et al (2009) SHOREmap simultaneous mapping and mutation identification by deep sequencing. Nat Methods 6:550–551
Segami S, Kono I, Ando T, Yano M, Kitano H, Miura K et al (2012) Small and round seed 5 gene encodes alpha-tubulin regulating seed cell elongation in rice. Rice 5:4
Shi Z, Rao Y, Xu J, Hu S, Fang Y, Yu H et al (2015) Characterization and cloning of SMALL GRAIN 4, a novel DWARF11 allele that affects brassinosteroid biosynthesis in rice. Sci Bull 60:905–915
Song XJ, Huang W, Shi M, Zhu MZ, Lin HX (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet 39:623–630
Sun Q, Wang K, Yoshimura A, Doi K (2002) Genetic differentiation for nuclear, mitochondrial and chloroplast genomes in common wild rice (Oryza rufipogon Griff.) and cultivated rice (Oryza sativa L.). Theor Appl Genet 104:1335–1345
Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C et al (2013) QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J 74:174–183
Takagi H, Tamiru M, Abe A, Yoshida K, Uemura A, Yaegashi H et al (2015) MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat Biotechnol 33:445–449
Tan L, Zhang P, Liu F, Wang G, Ye S, Zhu Z et al (2008) Quantitative trait loci underlying domestication- and yield-related traits in an Oryza sativa × Oryza rufipogon advanced backcross population. Genome 51:692–704
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Trick M, Adamski NM, Mugford SG, Jiang CC, Febrer M, Uauy C (2012) Combining SNP discovery from next-generation sequencing data with bulked segregant analysis (BSA) to fine-map genes in polyploid wheat. BMC Plant Biol 12:14
Tuinstra MR, Ejeta G, Goldsbrough PB (1997) Heterogeneous inbred family (HIF) analysis a method for developing near-isogenic lines that differ at quantitative trait loci. Theor Appl Genet 95:1005–1011
Unnevehr LJ, Duff B, Juliano BO (1992) Consumer demand for rice grain quality. International Rice Research Institute, Manila, and International Development Research Center, Ottawa
Varshney RK, Terauchi R, McCouch SR (2014) Harvesting the promising fruits of genomics: applying genome sequencing technologies to crop breeding. PLoS Biol 12:e1001883
Wang ZY, Second G, Tanksley SD (1992) Polymorphism and phylogenetic relationships among species in the genus Oryza as determined by analysis of nuclear RFLPs. Theor Appl Genet 83:565–581
Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X et al (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44:950–954
Wang S, Li S, Liu Q, Wu K, Zhang J, Wang S et al (2015a) The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat Genet 47:949–954
Wang Y, Xiong G, Hu J, Jiang L, Yu H, Xu J et al (2015b) Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat Genet 47:944–948
Wei X, Qiao WH, Chen YT, Wang RS, Cao LR, Zhang WX et al (2012) Domestication and geographic origin of Oryza sativa in China: insights from multilocus analysis of nucleotide variation of O. sativa and O. rufipogon. Mol Ecol 21:5073–5087
Weng J, Gu S, Wan X, Gao H, Guo T, Su N et al (2008) Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res 18:1199–1209
Xing Y, Zhang Q (2010) Genetic and molecular bases of rice yield. Annu Rev Plant Biol 61:421–442
Xu MR, Huang LY, Zhang F, Zhu LH, Zhou YL, Li ZK (2013) Genome-wide phylogenetic analysis of stress-activated protein kinase genes in rice (OsSAPKs) and expression profiling in response to Xanthomonas oryzae pv. oryzicola infection. Plant Mol Biol Rep 31:877–885
Yamamoto T, Kuboki Y, Lin SY et al (1998) Fine mapping of quantitative trait loci Hd-1, Hd-2 and Hd-3, controlling heading date of rice, as single Mendelian factors. Theor Appl Genet 97:37–44
Yamanaka N, Watanabe S, Toda K (2005) Fine mapping of the FT1 locus for soybean flowering time using a residual heterozygous line derived from a recombinant inbred line. Theor Appl Genet 110:634–639
Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P et al (2004) The wheat VRN2 gene is a flowering repressor downregulated by vernalization. Science 303:1640–1644
Yang Z, Huang D, Tang W, Zheng Y, Liang K, Cutler AJ et al (2013) Mapping of quantitative trait loci underlying cold tolerance in rice seedlings via high-throughput sequencing of pooled extremes. PLoS One 8:e68433
Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T et al (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12:2473–2483
Zheng KL, Huang N, Bennett J, and Khush GS (1995) PCR-based marker assisted selection in rice breeding. IRRI discussion paper series No. 12
Acknowledgements
We thank Shijia Liu, Liangming Chen, Xi Liu, and Yunlu Tian in Nanjing Agricultural University, China, for their assistance in field management at Nanjing experimental station. We thank Shelley Robison, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Funding
This study was supported by a Grant from the National Natural Science Foundation of China (No. 31471471), the National Key Research and Development Program of China (2016YFD0100101), and the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Science to Weihua Qiao.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical standards
The authors declare that this study complies with the current laws of the countries in which the experiments were performed.
Additional information
Communicated by Takuji Sasaki.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Qi, L., Ding, Y., Zheng, X. et al. Fine mapping and identification of a novel locus qGL12.2 control grain length in wild rice (Oryza rufipogon Griff.). Theor Appl Genet 131, 1497–1508 (2018). https://doi.org/10.1007/s00122-018-3093-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-018-3093-7