Abstract
Key message
Candidate genes associated with lignin and lodging traits were identified by combining phenotypic, genotypic, and gene expression data in B. napus.
Abstract
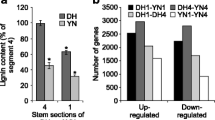
Brassica napus is one of the world’s most important oilseed crops, but its yield can be dramatically reduced by lodging, bending, and falling of its vertical stems. Lignin has been shown to contribute to stem mechanical strength. In this study, we found that the syringyl/guaiacyl (S/G) monolignol ratio exhibits a significant negative correlation with disease and lodging resistance. A total of 92 and 50 SNP and SSR loci, respectively, were found to be significantly associated with five traits, breaking force, breaking strength, lodging coefficient, acid detergent lignin content, and the S/G monolignol ratio using GWAS. To identify novel genes involved in lignin biosynthesis, transcriptome sequencing of high- (H) and low (L)-ADL content accessions was performed. The up-regulated genes were mainly involved in glycoside catabolic processes (especially glucosinolate catabolism) and cell wall biogenesis, while down-regulated genes were involved in glucosinolate biosynthesis, indicating that crosstalk exists between glucosinolate metabolic processes and lignin biosynthesis. Integrating this differential expression with the GWAS analysis, we identified four candidate genes regulating lignin, including glycosyl hydrolase (BnaA01g00480D), CYT1 (BnaA04g22820D), and two encoding transcription factors, SHINE1 (ERF family) and DAR6 (LIM family). This study provides insight into the genetic control of lodging and lignin in B. napus.





Similar content being viewed by others
References
Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A (2004) The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16:2463–2480
Ambavaram MM, Krishnan A, Trijatmiko KR, Pereira A (2011) Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiol 155:916–931
Barriere Y, Courtial A, Soler M, Grima-Pettenati J (2015) Toward the identification of genes underlying maize QTLs for lignin content, focusing on colocalizations with lignin biosynthetic genes and their regulatory MYB and NAC transcription factors. Mol Breed 35:87
Berry PM, Sterling M, Spink JH, Baker CJ, Sylvester-Bradley R, Mooney SJ, Tams AR, Ennos AR (2004) Understanding and reducing lodging in cereals. Adv Agron 84:217–271
Board J (2001) Reduced lodging for soybean in low plant population is related to light quality. Crop Sci 41:379–384
Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54:519–546
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Cappa EP, El-Kassaby YA, Garcia MN, Acuna C, Borralho NM, Grattapaglia D, Marcucci Poltri SN (2013) Impacts of population structure and analytical models in genome-wide association studies of complex traits in forest trees: a case study in Eucalyptus globulus. PLoS One 8:e81267
Capron A, Chang XF, Hall H, Ellis B, Beatson RP, Berleth T (2013) Identification of quantitative trait loci controlling fibre length and lignin content in Arabidopsis thaliana stems. J Exp Bot 64:185–197
Chavigneau H, Goué N, Courtial A, Jouanin L, Reymond M, Méchin V, Barrière Y (2012) QTL for floral stem lignin content and degradability in three recombinant inbred line (RIL) progenies of Arabidopsis thaliana and search for candidate genes involved in cell wall biosynthesis and degradability. OJGen 2:7–30
Chen HF, Shan ZH, Sha AH, Wu BD, Yang ZL, Chen SL, Zhou R, Zhou XN (2011) Quantitative trait loci analysis of stem strength and related traits in soybean. Euphytica 179:485–497
Chhatre VE, Emerson KJ (2017) StrAuto: automation and parallelization of Structure analysis. BMC Bioinformatics 18:192
Davidson J, Phillips M (1930) Lignin as a possible factor in lodging of cereals. Science 72:401–402
Dixit S, Grondin A, Lee CR, Henry A, Olds TM, Kumar A (2015) Understanding rice adaptation to varying agro-ecosystems: trait interactions and quantitative trait loci. BMC Genet 16:86
Earl DA, Vonholdt BM (2012) Structure Harvester: a website and program for visualizing Structure output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Escamilla-Trevino LL, Chen W, Card ML, Shih MC, Cheng CL, Poulton JE (2006) Arabidopsis thaliana beta-glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides. Phytochemistry 67:1651–1660
Flintham JE, Borner A, Worland AJ, Gale MD (1997) Optimizing wheat grain yield: effects of Rht (gibberellin-insensitive) dwarfing genes. J Agr Sci 128:11–25
Foulkes MJ, Slafer GA, Davies WJ, Berry PM, Sylvester-Bradley R, Martre P, Calderini DF, Griffiths S, Reynolds MP (2011) Raising yield potential of wheat. III. Optimizing partitioning to grain while maintaining lodging resistance. J Exp Bot 62:469–486
Gigolashvili T, Yatusevich R, Berger B, Muller C, Flugge UI (2007) The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant Journal 51:247–261
Guragain YN, Herrera AI, Vadlani PV, Prakash O (2015) Lignins of bioenergy crops: a review? Nat Prod Commun 10:201–208
Gyawali S, Harrington M, Durkin J, Horner K, Parkin IAP, Hegedus DD, Bekkaoui D, Buchwaldt L (2016) Microsatellite markers used for genome-wide association mapping of partial resistance to Sclerotinia sclerotiorum in a world collection of Brassica napus. Mol Breed 36:72
Hai L, Guo HH, Xiao SH, Jiang GL, Zhang XY, Yan CS, Xin ZY, Jia JZ (2005) Quantitative trait loci (QTL) of stem strength and related traits in a doubled-haploid population of wheat (Triticum aestivum L.). Euphytica 141:1–9
Hardy OJ, Vekemans X (2002) SPAGEDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Hemm MR, Ruegger MO, Chapple C (2003) The Arabidopsis ref2 mutant is defective in the gene encoding CYP83A1 and shows both phenylpropanoid and glucosinolate phenotypes. Plant Cell 15:179–194
Huang S, Cerny RE, Bhat DS, Brown SM (2001) Cloning of an Arabidopsis patatin-like gene, STURDY, by activation T-DNA tagging. Plant Physiol 125:573–584
Islam N, Evans EJ (1994) Influence of lodging and nitrogen rate on the yield and yield attributes of oilseed rape (Brassica napus L.). Theor Appl Genet 88:530–534
Kannangara R, Branigan C, Liu Y, Penfield T, Rao V, Mouille G, Hofte H, Pauly M, Riechmann JL, Broun P (2007) The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell 19:1278–1294
Kawaoka A, Kaothien P, Yoshida K, Endo S, Yamada K, Ebinuma H (2000) Functional analysis of tobacco LIM protein Ntlim1 involved in lignin biosynthesis. Plant J 22:289–301
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578
Turner SD (2014) qqman: an R package for visualizing GWAS results using Q–Q and manhattan plots. bioRxiv 005165
Kim JI, Dolan WL, Anderson NA, Chapple C (2015) Indole glucosinolate biosynthesis limits phenylpropanoid accumulation in Arabidopsis thaliana. Plant Cell 27:1529–1546
Kishimoto T, Chiba W, Saito K, Fukushima K, Uraki Y, Ubukata M (2010) Influence of syringyl to guaiacyl ratio on the structure of natural and synthetic lignins. J Agric Food Chem 58:895–901
Lex J, Ahlemeyer J, Friedt W, Ordon F (2014) Genome-wide association studies of agronomic and quality traits in a set of German winter barley (Hordeum vulgare L.) cultivars using Diversity Arrays Technology (DArT). J Appl Genet 55:295–305
Li F, Chen B, Xu K, Wu J, Song W, Bancroft I, Harper AL, Trick M, Liu S, Gao G, Wang N, Yan G, Qiao J, Li J, Li H, Xiao X, Zhang T, Wu X (2014a) Genome-wide association study dissects the genetic architecture of seed weight and seed quality in rapeseed (Brassica napus L.). DNA Res 21:355–367
Li Y, Gu H, Qi C (2014b) QTL mapping for lodging resistance and its related traits by RIL population of Brassica napus L. Chin J Oil Crop Sci 36:10–17
Li FC, Zhang ML, Guo K, Hu Z, Zhang R, Feng YQ, Yi XY, Zou WH, Wang LQ, Wu CY, Tian JS, Lu TG, Xie GS, Peng LC (2015) High-level hemicellulosic arabinose predominately affects lignocellulose crystallinity for genetically enhancing both plant lodging resistance and biomass enzymatic digestibility in rice mutants. Plant Biotechnol J 13:514–525
Liepman AH, Wightman R, Geshi N, Turner SR, Scheller HV (2010) Arabidopsis—a powerful model system for plant cell wall research. Plant J 61:1107–1121
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Liu T, Guan C, Xiao J, Liang Y, Lei D, Wang Y (2007) Relation between physico-chemical properties of stem and lodging and evaluation of lodging resistance in rapeseed (Brassica napus L.). J Henan Agric Sci 12:40–42
Liu L, Qu C, Wittkop B, Yi B, Xiao Y, He Y, Snowdon RJ, Li J (2013) A high-density SNP map for accurate mapping of seed fibre QTL in Brassica napus L. PLoS One 8:e83052
Lukowitz W, Nickle TC, Meinke DW, Last RL, Conklin PL, Somerville CR (2001) Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc Natl Acad Sci USA 98:2262–2267
Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21:3448–3449
Miralles DJ, Slafer GA (1995) Individual grain weight responses to genetic reduction in culm length in wheat as affected by source-sink manipulations. Field Crop Res 43:55–66
Nickle TC, Meinke DW (1998) A cytokinesis-defective mutant of Arabidopsis (cyt1) characterized by embryonic lethality, incomplete cell walls, and excessive callose accumulation. Plant J 15:321–332
Ohman D, Demedts B, Kumar M, Gerber L, Gorzsas A, Goeminne G, Hedenstrom M, Ellis B, Boerjan W, Sundberg B (2013) MYB103 is required for FERULATE-5-HYDROXYLASE expression and syringyl lignin biosynthesis in Arabidopsis stems. Plant J 73:63–76
Ookawa T, Hobo T, Yano M, Murata K, Ando T, Miura H, Asano K, Ochiai Y, Ikeda M, Nishitani R, Ebitani T, Ozaki H, Angeles ER, Hirasawa T, Matsuoka M (2010) New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat Commun 1:132
Pei Y, Li Y, Zhang Y, Yu C, Fu T, Zou J, Tu Y, Peng L, Chen P (2016) G-lignin and hemicellulosic monosaccharides distinctively affect biomass digestibility in rapeseed. Biores Technol 203:325–333
Peiffer JA, Flint-Garcia SA, De Leon N, McMullen MD, Kaeppler SM, Buckler ES (2013) The genetic architecture of maize stalk strength. PLoS One 8:e67066
Pena MJ, Zhong RQ, Zhou GK, Richardson EA, O’Neill MA, Darvill AG, York WS, Ye ZH (2007) Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19:549–563
Peng X (2012) The selection of lodging indicators and mapping QTL for lodging in Brassica napus L. Master’s thesis Southwest University, Chongqing, China
Qu CM, Li SM, Duan XJ, Fan JH, Jia LD, Zhao HY, Lu K, Li JN, Xu XF, Wang R (2015) Identification of candidate genes for seed glucosinolate content using association mapping in Brassica napus L. Genes 6:1215–1229
Robinson AR, Mansfield SD (2009) Rapid analysis of poplar lignin monomer composition by a streamlined thioacidolysis procedure and near-infrared reflectance-based prediction modeling. Plant J 58:706–714
Rode J, Ahlemeyer J, Friedt W, Ordon F (2012) Identification of marker-trait associations in the German winter barley breeding gene pool (Hordeum vulgare L.). Mol Breed 30:831–843
Rogers LA, Campbell MM (2004) The genetic control of lignin deposition during plant growth and development. New Phytol 164:17–30
Rohlf FJ (2000) NTSYS-pc: numerical taxonomy and multivariate analysis system: version 2.1. Exeter Software
Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27:431–432
Soto-Cerda BJ, Duguid S, Booker H, Rowland G, Diederichsen A, Cloutier S (2014) Genomic regions underlying agronomic traits in linseed (Linum usitatissimum L.) as revealed by association mapping. J Integr Plant Biol 56:75–87
Vanholme R, Morreel K, Darrah C, Oyarce P, Grabber JH, Ralph J, Boerjan W (2012a) Metabolic engineering of novel lignin in biomass crops. New Phytol 196:978–1000
Vanholme R, Storme V, Vanholme B, Sundin L, Christensen JH, Goeminne G, Halpin C, Rohde A, Morreel K, Boerjan W (2012b) A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 24:3506–3529
Vanholme R, Cesarino I, Rataj K, Xiao YG, Sundin L, Goeminne G, Kim H, Cross J, Morreel K, Araujo P, Welsh L, Haustraete J, McClellan C, Vanholme B, Ralph J, Simpson GG, Halpin C, Boerjan W (2013) Caffeoyl shikimate esterase (CSE) Is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341:1103–1106
Vansoest PJ, Robertson JB, Lewis BA (1991) Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci 74:3583–3597
Wang J, Jian H, Wei L, Qu C, Xu X, Lu K, Qian W, Li J, Li M, Liu L (2015) Genome-wide analysis of seed acid detergent lignin (ADL) and hull content in rapeseed (Brassica napus L.). PLoS One 10:e0145045
Wegrzyn JL, Eckert AJ, Choi M, Lee JM, Stanton BJ, Sykes R, Davis MF, Tsai CJ, Neale DB (2010) Association genetics of traits controlling lignin and cellulose biosynthesis in black cottonwood (Populus trichocarpa, Salicaceae) secondary xylem. New Phytol 188:515–532
Wei LJ, Jian HJ, Lu K, Filardo F, Yin NW, Liu LZ, Qu CM, Li W, Du H, Li JN (2016) Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant Biotechnol J 14:1368–1380
Xie JK, Kong XL, Chen J, Hu BL, Wen PA, Zhuang JY, Bao JS (2011) Mapping of quantitative trait loci for fiber and lignin contents from an interspecific cross Oryza sativaxOryza rufipogon. J Zhejiang Univ-Sc B 12:518–526
Yano K, Ookawa T, Aya K, Ochiai Y, Hirasawa T, Ebitani T, Takarada T, Yano M, Yamamoto T, Fukuoka S, Wu J, Ando T, Ordonio RL, Hirano K, Matsuoka M (2015) Isolation of a novel lodging resistance QTL gene involved in strigolactone signaling and its pyramiding with a QTL gene involved in another mechanism. Mol Plant 8:303–314
Zeng J, Singh D, Gao D, Chen S (2014) Effects of lignin modification on wheat straw cell wall deconstruction by Phanerochaete chrysosporium. Biotechnol Biofuels 7:161
Zhong RQ, Burk DH, Morrison WH, Ye ZH (2002) A kinesin-like protein is essential for oriented deposition of cellulose microfibrils and cell wall strength. Plant Cell 14:3101–3117
Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20:2763–2782
Ziebell A, Gracom K, Katahira R, Chen F, Pu Y, Ragauskas A, Dixon RA, Davis M (2010) Increase in 4-coumaryl alcohol units during lignification in alfalfa (Medicago sativa) alters the extractability and molecular weight of lignin. J Biol Chem 285:38961–38968
Acknowledgments
This work was supported by the National Science and Technology (2013BAD01B03-12) and the National High Technology Research and Development Plan (2016YFD0101007), 863 project (2011AA10A104 and 2013AA102602), the National Natural Science Foundation of China (31171619, 31371655, and 31571701 and U1302266), the Earmarked Fund for Modern Agro-industry Technology Research System (CARS-13), the “111″ Project (B12006), and the postdoctoral programme (BX201700201). Thanks to the China Scholarship Council (CSC) for its support.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declared no conflict of interest in the authorship and publication of this document.
Additional information
Communicated by Carlos F. Quiros.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wei, L., Jian, H., Lu, K. et al. Genetic and transcriptomic analyses of lignin- and lodging-related traits in Brassica napus . Theor Appl Genet 130, 1961–1973 (2017). https://doi.org/10.1007/s00122-017-2937-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-017-2937-x



