Abstract
Key message
C genome chromosome substitution lines of B. juncea constitute a key genetic resource for increased genetic diversity and hybrid performance.
Abstract
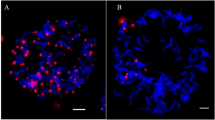
C genome chromosome substitution lines were found in the progenies of derived B. juncea (2n = 36; AABB), synthesized through hybridization between B. napus and B. carinata. These were originally recognized based on the morphology and genomic in situ hybridization. Genotyping using the Brassica Illumina 60K Infinium SNP array confirmed the presence of C genome chromosomes in a large number of derived B. juncea genotypes. Three whole chromosome substitutions and 13 major C genome fragment substitutions were identified. Fragment substitutions were primarily terminal, but intercalary substitution(s) involving chromosome C1 and C2 were identified in three genotypes. The size of substituted C genome fragments varied from 0.04 Mbp (C1) to 64.85 Mbp (C3). In terms of proportions, these ranged from 0.10 % (C1) to 100 % (C1, C3 and C7) of the substituted chromosome. SSR genotyping with B genome specific primers suggested that substituting C genome chromosome(s) are likely to have replaced B genome chromosome(s). C1 was the most common substituting chromosome while substituted B chromosome seemed random. Study of the phenotypic traits underlined the importance of the substitution lines (especially of chromosome C1) for conferring superior trait performance (main shoot length and pods on the main shoot). High heterosis was also indicated in hybrid combinations of substitution lines with natural B. juncea. These substitution genotypes constituted a valuable resource for targeted gene transfer and QTL identification.









Similar content being viewed by others
References
Atri C, Kumar B, Kumar H, Kumar S, Sharma S, Banga SS (2012) Development and characterization of Brassica juncea–fruticulosa introgression lines exhibiting resistance to mustard aphid (Lipaphis erysimi Kalt). BMC Genet 13:104
Atri C, Kaur B, Sharma S, Gandhi N, Verma H, Goyal A, Banga SS (2016) Substituting nuclear genome of Brassica juncea (L.) Czern and Coss. In: Cytoplasmic background of Brassica fruticulosa results in cytoplasmic male sterility. Euphytica. doi:10.1007/s10681-015-1628-4
Banga SS (1988) C-genome chromosome substitution lines in Brassica juncea (L.) Czern and Coss. Genetica 77:81–84
Banga SS, Banga SK (2009) Crop improvement strategies in rapeseed-mustard. In: Hegde DM (ed) Vegetable oils scenario: approaches to meet the growing demands. ISOR, Hyderabad, pp 13–35
Banga SS, Bhaskar PB, Ahuja I (2003) Synthesis of intergeneric hybrids and establishment of genomic affinity between Diplotaxis catholica and crop Brassica species. Theor Appl Genet 106:1244–1247
Banuelos GS, Dhillon SK, Banga SS (2013) Oilseed Brassicas. In: Singh BP (ed) Biofuel crops: production, physiology and genetics. Centre for Agricultural Biosciences International, Egham, pp 339–368
Chauhan JS, Singh KH, Singh VV, Kumar S (2011) Hundred years of rapeseed-mustard breeding in India: accomplishments and future strategies. Indian J Agr Sci 81:1093–1109
Chen BY, Cheng BF, Jorgensen RB, Heneen WK (1997) Production and cytogenetics of Brassica campestris-alboglabra chromosome addition lines. Theor Appl Genet 94:633–640
Cheng F, Mandakova T, Wu J, Xie Q, Lysak MA, Wang X (2013) Deciphering the diploid ancestral genome of the mesohexaploid Brassica rapa. Plant Cell 25:1541–1544
Cowling W (2007) Genetic diversity in Australian canola and implications for crop breeding for changing future environments. Field Crop Res 104:103–111
Delourme R, Eber F, Cheavre AM (1989) Intergeneric hybridization of Diplotaxis erucoides and Brassica napus I. Cytogenetic analysis of F1 and BC1 progeny. Euphytica 41:123–128
Ding G, Zhao Z, Liao Y et al (2012) Quantitative trait loci for seed yield and yield-related traits, and their responses to reduced phosphorus supply in Brassica napus. Ann Bot 109:747–759
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Garg H, Atri C, Sandhu PS, Kaur B, Renton M, Banga SK, Singh H, Singh C, Barbetti MJ, Banga SS (2010) High level of resistance to Sclerotinia sclerotiorum in introgression lines derived from hybridization between wild crucifers and the crop Brassica species B. napus and B. juncea. Field Crop Res 117:51–58
Gupta M, Gupta S, Kumar H, Kumar N, Banga SS (2015) Population structure and breeding value of a new type of Brassica juncea created by combining A and B genomes from related allotetraploids. Theor Appl Genet 128:221–234
Jahier J, Chevre A, Tanguy A, Eber F (1989) Extraction of disomic addition lines of Brassica napus–B. nigra. Genome 32:408–413
Janeja HS, Banga SK, Bhaskar PB, Banga SS (2003) Alloplasmic male sterile Brassica napus with Enarthrocarpus lyratus cytoplasm: introgression and molecular mapping of an E. lyratus chromosome segment carrying a fertility restoring gene. Genome 46(5):792–797
Kaur P, Banga S, Kumar N, Gupta S, Akhatar J, Banga SS (2014) Polyphyletic origin of Brassica juncea with B. rapa and B. nigra (Brassicaceae) participating as cytoplasm donor parents in independent hybridization events. Am J Bot 101(7):1157–1166
Krzywinski MI, Schein JE, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19(9):1639–1645
Kumar S, Atri C, Sangha MK, Banga SS (2011) Screening of wild crucifers for resistance to mustard aphid, Lipaphis erysimi (Kaltenbach) and attempt at introgression of resistance gene(s) from Brassica fruticulosa to Brassica juncea. Euphytica 179:461–470
Mason AS, Huteau V, Eber F, Coriton O, Yan G, Nelson M, Cowling WA, Chevre AM (2010) Genome structure affects the rate of autosyndesis and allosyndesis in AABC, BBAC and CCAB Brassica interspecific hybrids. Chromosome Res 18:655–666
Mason AS, Batley J, Bayer PE, Hayward A, Cowling WA, Nelson MN (2014a) High-resolution molecular karyotyping uncovers pairing between ancestrally related Brassica chromosomes. New Phytol 202(3):964–974
Mason AS, Nelson MN, Takahira J, Cowling WA, Alves GM, Chaudhuri A, Chen N, Ragu ME, Dalton-Morgan J, Coriton O, Huteau H, Eber F, Chèvre AM, Batley J (2014b) The fate of chromosomes and alleles in an allohexaploid Brassica population. Genetics 197:273–283
Mason AS, Zhang J, Tollenaere R, Vasquez Teuber P, Dalton-Morgan J, Hu L et al (2015) High-throughput genotyping for species identification and diversity assessment in germplasm collections. Mol Ecol Resour. doi:10.1111/1755-0998.12379
Nadeau JH, Singer JB, Matin A, Lander ES (2000) Analysing complex genetic traits with chromosome substitution strains. Nat Genet 24:221–225
Parkin IAP, Koh C, Tang H, Robinson SJ, Kagale S, Clarke WE, Town CD, Nixon J, Krishnakumar K et al (2014) Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol 15:R77
Prakash S, Bhat SR, Qurios CF, Kirit PB, Chopra VL (2009) Brassica and its close allies: cytogenetics and evolution. Plant Breed Rev 31:21–187
Raman H, Raman R, Kilian A, Detering F, Carling J, Coombes N, Diffey S, Kadkol G, Edwards D, McCully M, Kumar P, Parkin IAP, Batley J et al (2014) Genome-wide delineation of natural variation for pod shatter resistance in Brassica napus. PLoS ONE 9:e101673
Schwarzacher T, Heslop-Harrison JS (2000) Practical in situ hybridization. BIOS Scientific Publishers, Oxford
Schwarzacher T, Anamthawat-Jónsson K, Harrison GE, Islam AKMR, Jia JZ, King IP et al (1992) Genomic in situ hybridization to identify alien chromosomes and chromosome segments in wheat. Theor Appl Genet 84:778–786
Shi J, Li R, Qiu D, Jiang C, Long Y, Morgan C, Bancroft I, Zhao J, Meng J (2009) Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus. Genetics 182:3851–3861
Truco MJ, Hu J, Sadowski J, Quiros CF (1996) Inter- and intragenomic homology of the Brassica genomes: implications for their origin and evolution. Theor Appl Genet 93:1225–1233
Van Berloo R (2008) GGT 2.0: versatile software for visualization and analysis of genetic data. J Hered 99:232–236
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F, Huang S, Li X, Hua W, Wang J, Wang X, Freeling M, Pires JC, Paterson AH, Chalhoub B, Wang B, Hayward A, Sharpe AG, Park BS, Weisshaar B, Liu B, Li B, Liu B, Tong C, Song C, Duran C et al (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Gen 43:1035–1039
Warwick SI (2011) Brassicaceae in agriculture. In: Schmidt R, Bancroft I (eds) Genetics and genomics of the Brassicaceae. Plant genetics and genomics: crops and models. Springer, New York, pp 33–65
Zamir D (2001) Improving plant breeding with exotic genetic libraries. Nat Rev Genet 2:983–989
Zhou QH, Fu DH, Mason AS, Zeng YJ, Zhao CX, Huan YJ (2014) In silico integration of quantitative trait loci for seed yield and yield-related traits in Brassica napus. Mol Breed 33:881–894
Acknowledgments
Research was carried out with the financial assistance from Department of Biotechnology, Government of India in form of Centre of Excellence and Innovation in Biotechnology “Germplasm enhancement for crop architecture and defensive traits in Brassica juncea l. czern. and coss.” Surinder Banga also acknowledges salary support from Indian Council of Agricultural Research. Jacqueline Batley is supported by an Australian Research Council Future Fellowship (FT130100604). Annaliese Mason acknowledges salary support from an Australian Research Council Discovery Early Career Researcher Award (DE120100668) and a Deutsche Forschungsgemeinschaft Emmy Noether award (MA 6473/1-1).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by P. Heslop-Harrison.
Rights and permissions
About this article
Cite this article
Gupta, M., Mason, A.S., Batley, J. et al. Molecular-cytogenetic characterization of C-genome chromosome substitution lines in Brassica juncea (L.) Czern and Coss.. Theor Appl Genet 129, 1153–1166 (2016). https://doi.org/10.1007/s00122-016-2692-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-016-2692-4