Abstract
Key message
Genomic selection models can be trained using historical data and filtering genotypes based on phenotyping intensity and reliability criterion are able to increase the prediction ability.
Abstract
We implemented genomic selection based on a large commercial population incorporating 2325 European winter wheat lines. Our objectives were (1) to study whether modeling epistasis besides additive genetic effects results in enhancement on prediction ability of genomic selection, (2) to assess prediction ability when training population comprised historical or less-intensively phenotyped lines, and (3) to explore the prediction ability in subpopulations selected based on the reliability criterion. We found a 5 % increase in prediction ability when shifting from additive to additive plus epistatic effects models. In addition, only a marginal loss from 0.65 to 0.50 in accuracy was observed using the data collected from 1 year to predict genotypes of the following year, revealing that stable genomic selection models can be accurately calibrated to predict subsequent breeding stages. Moreover, prediction ability was maximized when the genotypes evaluated in a single location were excluded from the training set but subsequently decreased again when the phenotyping intensity was increased above two locations, suggesting that the update of the training population should be performed considering all the selected genotypes but excluding those evaluated in a single location. The genomic prediction ability was substantially higher in subpopulations selected based on the reliability criterion, indicating that phenotypic selection for highly reliable individuals could be directly replaced by applying genomic selection to them. We empirically conclude that there is a high potential to assist commercial wheat breeding programs employing genomic selection approaches.





Similar content being viewed by others
References
Akdemir D, Sanchez JI, Jannink J-L (2015) Optimization of genomic selection training populations with a genetic algorithm. Genet Sel Evol 47:38
Asoro FG, Newell MA, Beavis WD, Scott MP, Jannink J-L (2011) Accuracy and training population design for genomic selection on quantitative traits in elite North American oats. Plant Genome 4:132–144
Boligon A, Long N, Albuquerque LGd, Weigel K, Gianola D, Rosa G (2012) Comparison of selective genotyping strategies for prediction of breeding values in a population undergoing selection. J Anim Sci 90:4716–4722
Burgueño J, de los Campos G, Weigel K, Crossa J (2012) Genomic prediction of breeding values when modeling genotype × environment interaction using pedigree and dense molecular markers. Crop Sci 52:707–719
Cai X, Huang A, Xu S (2011) Fast empirical Bayesian LASSO for multiple quantitative trait locus mapping. BMC Bioinform 12:211
Cavanagh CR, Chao S, Wang S, Huang BE, Stephen S, Kiani S, Forrest K, Saintenac C, Brown-Guedira GL, Akhunova A (2013) Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc Natl Acad Sci 110:8057–8062
Crossa J, de los Campos G, Pérez P, Gianola D, Burgueno J, Araus JL, Makumbi D, Singh RP, Dreisigacker S, Yan J (2010) Prediction of genetic values of quantitative traits in plant breeding using pedigree and molecular markers. Genetics 186:713–724
Dawson JC, Endelman JB, Heslot N, Crossa J, Poland J, Dreisigacker S, Manès Y, Sorrells ME, Jannink J-L (2013) The use of unbalanced historical data for genomic selection in an international wheat breeding program. Field Crops Res 154:12–22
de los Campos G, Gianola D, Rosa GJ, Weigel KA, Crossa J (2010) Semi-parametric genomic-enabled prediction of genetic values using reproducing kernel Hilbert spaces methods. Genet Res 92:295–308
Falconer DS (1960) Introduction to quantitative genetics. DS Falconer
Garrick DJ, Taylor JF, Fernando RL (2009) Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet Sel Evol 41
Gianola D, van Kaam JB (2008) Reproducing kernel Hilbert spaces regression methods for genomic assisted prediction of quantitative traits. Genetics 178:2289–2303
Gianola D, Fernando RL, Stella A (2006) Genomic-assisted prediction of genetic value with semiparametric procedures. Genetics 173:1761–1776
Gilmour AR, Gogel B, Cullis B, Thompson R, Butler D (2009) ASReml user guide release 3.0. VSN International Ltd, Hemel Hempstead, UK
Goldringer I, Brabant P, Gallais A (1997) Estimation of additive and epistatic genetic variances for agronomic traits in a population of doubled-haploid lines of wheat. Heredity 79:60–71
González-Camacho J, de Los Campos G, Pérez P, Gianola D, Cairns J, Mahuku G, Babu R, Crossa J (2012) Genome-enabled prediction of genetic values using radial basis function neural networks. Theor Appl Genet 125:759–771
Gower JC (1966) Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53:325–338
Guo Z, Tucker DM, Basten CJ, Gandhi H, Ersoz E, Guo B, Xu Z, Wang D, Gay G (2014) The impact of population structure on genomic prediction in stratified populations. Theor Appl Genet 127:749–762
Habier D, Fernando R, Dekkers J (2007) The impact of genetic relationship information on genome-assisted breeding values. Genetics 177:2389–2397
Habier D, Fernando RL, Kizilkaya K, Garrick DJ (2011) Extension of the Bayesian alphabet for genomic selection. BMC Bioinform 12:186
Hayes B, Bowman P, Chamberlain A, Goddard M (2009a) Invited review: genomic selection in dairy cattle: Progress and challenges. J Dairy Sci 92:433–443
Hayes B, Bowman P, Chamberlain A, Verbyla K, Goddard M (2009b) Accuracy of genomic breeding values in multi-breed dairy cattle populations. Genetics Selection Evolution 41:51
Heffner EL, Sorrells ME, Jannink J-L (2009) Genomic selection for crop improvement. Crop Sci 49:1–12
Heffner EL, Lorenz AJ, Jannink J-L, Sorrells ME (2010) Plant breeding with genomic selection: gain per unit time and cost. Crop Sci 50:1681–1690
Henderson CR (1973) Sire evaluation and genetic trends. J Anim Sci 1973:10–41
Henderson CR (1975) Best linear unbiased estimation and prediction under a selection model. Biometrics: 423–447
Henderson CR (1985) Best linear unbiased prediction of nonadditive genetic merits. J Anim Sci 60:111–117
Heslot N, Yang H-P, Sorrells ME, Jannink J-L (2012) Genomic selection in plant breeding: a comparison of models. Crop Sci 52:146–160
Howie BN, Donnelly P, Marchini J (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5:e1000529
Isidro J, Jannink J-L, Akdemir D, Poland J, Heslot N, Sorrells ME (2015) Training set optimization under population structure in genomic selection. Theor Appl Genet 128:145–158
Jannink J-L, Lorenz AJ, Iwata H (2010) Genomic selection in plant breeding: from theory to practice. Briefings in functional genomics 9:166–177
Jiang Y, Reif JC (2015) Modeling epistasis in genomic selection. Genetics 201:759–768
Jiménez-Montero J, Gonzalez-Recio O, Alenda R (2012) Genotyping strategies for genomic selection in small dairy cattle populations. Animal 6:1216–1224
Liu Z, Seefried FR, Reinhardt F, Rensing S, Thaller G, Reents R (2011) Impacts of both reference population size and inclusion of a residual polygenic effect on the accuracy of genomic prediction. Genet Sel Evol 43
Longin CFH, Mi X, Würschum T (2015) Genomic selection in wheat: optimum allocation of test resources and comparison of breeding strategies for line and hybrid breeding. Theor Appl Genet 128:1297–1306
Lopez-Cruz M, Crossa J, Bonnett D, Dreisigacker S, Poland J, Jannink J-L, Singh RP, Autrique E, de los Campos G (2015) Increased prediction accuracy in wheat breeding trials using a marker × environment interaction genomic selection model. G3: Genes| Genomes| Genetics:g3. 114.016097
Lorenzana RE, Bernardo R (2009) Accuracy of genotypic value predictions for marker-based selection in biparental plant populations. Theor Appl Genet 120:151–161
Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Mirdita V, He S, Zhao Y, Korzun V, Bothe R, Ebmeyer E, Reif JC, Jiang Y (2015) Potential and limits of whole genome prediction of resistance to Fusarium head blight and Septoria tritici blotch in a vast Central European elite winter wheat population. Theor Appl Genet. doi:10.1007/s00122-015-2602-1
Möhring J, Piepho H-P (2009) Comparison of weighting in two-stage analysis of plant breeding trials. Crop Sci 49:1977–1988
Morota G, Gianola D (2014) Kernel-based whole-genome prediction of complex traits: a review. Front genet 5:363
Ostersen T, Christensen OF, Henryon M, Nielsen B, Su G, Madsen P (2011) Deregressed EBV as the response variable yield more reliable genomic predictions than traditional EBV in pure-bred pigs. Genet Sel Evol 43:38
Pérez P, de los Campos G (2014) Genome-wide regression and prediction with the BGLR statistical package. Genetics 198:483–495
Poland J, Endelman J, Dawson J, Rutkoski J, Wu S, Manes Y, Dreisigacker S, Crossa J, Sánchez-Villeda H, Sorrells M (2012) Genomic selection in wheat breeding using genotyping-by-sequencing. Plant Genome 5:103–113
Ray DK, Gerber JS, MacDonald GK, West PC (2015) Climate variation explains a third of global crop yield variability. Nat commun 6:5989
R Core Team (2014) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org/
Rincent R, Laloë D, Nicolas S, Altmann T, Brunel D, Revilla P, Rodriguez VM, Moreno-Gonzalez J, Melchinger A, Bauer E (2012) Maximizing the reliability of genomic selection by optimizing the calibration set of reference individuals: comparison of methods in two diverse groups of maize in breds (Zea mays L.). Genetics 192:715–728
Rogers JS (1972) Measures of genetic similarity and genetic distance. Stud genet 7:145–153
Rutkoski J, Benson J, Jia Y, Brown-Guedira G, Jannink JL, Sorrells ME (2012) Evaluation of genomic prediction methods for Fusarium head blight resistance in wheat. Plant Genome 5:51–61
Rutkoski J, Singh R, Huerta-Espino J, Bhavani S, Poland J, Jannink J, Sorrells M (2015) Efficient use of historical data for genomic selection: a case study of stem rust resistance in wheat. Plant Genome. doi:10.3835/plantgenome2014.09.0046
Sallam A, Endelman J, Jannink J-L, Smith K (2015) Assessing genomic selection prediction accuracy in a dynamic barley breeding population. Plant Genome. doi:10.3835/plantgenome2014.05.0020
Schulz-Streeck T, Ogutu JO, Piepho H-P (2013) Comparisons of single-stage and two-stage approaches to genomic selection. Theor Appl Genet 126:69–82
Utz H, Laidig F (1989) Genetic and environmental variability of yields in the official FRG variety performance tests. Biul Oceny Odmian:21–22
VanRaden P (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91:4414–4423
VanRaden P, Van Tassell C, Wiggans G, Sonstegard T, Schnabel R, Taylor J, Schenkel F (2009) Invited review: reliability of genomic predictions for North American Holstein bulls. J Dairy Sci 92:16–24
Wang D, El-Basyoni IS, Baenziger PS, Crossa J, Eskridge K, Dweikat I (2012) Prediction of genetic values of quantitative traits with epistatic effects in plant breeding populations. Heredity 109:313–319
Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, Maccaferri M, Salvi S, Milner SG, Cattivelli L (2014) Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796
Weber K, Thallman R, Keele J, Snelling W, Bennett G, Smith T, McDaneld T, Allan M, Van Eenennaam A, Kuehn L (2012) Accuracy of genomic breeding values in multibreed beef cattle populations derived from deregressed breeding values and phenotypes. J Anim Sci 90:4177–4190
Whittaker JC, Thompson R, Denham MC (2000) Marker-assisted selection using ridge regression. Genet Res 75:249–252
Windhausen VS, Atlin GN, Hickey JM, Crossa J, Jannink J-L, Sorrells ME, Raman B, Cairns JE, Tarekegne A, Semagn K (2012) Effectiveness of genomic prediction of maize hybrid performance in different breeding populations and environments. G3: genes| Genomes|. Genetics 2:1427–1436
Wittenburg D, Melzer N, Reinsch N (2011) Including non-additive genetic effects in Bayesian methods for the prediction of genetic values based on genome-wide markers. BMC Genet 12:74
Xu S (2007) An empirical Bayes method for estimating epistatic effects of quantitative trait loci. Biometrics 63:513–521
Zhao Y, Gowda M, Longin FH, Würschum T, Ranc N, Reif JC (2012) Impact of selective genotyping in the training population on accuracy and bias of genomic selection. Theor Appl Genet 125:707–713
Zhong S, Dekkers JC, Fernando RL, Jannink J-L (2009) Factors affecting accuracy from genomic selection in populations derived from multiple inbred lines: a barley case study. Genetics 182:355–364
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors agree that there are not conflicts of interest to be reported.
Additional information
Communicated by H. Iwata.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2015_2655_MOESM1_ESM.jpg
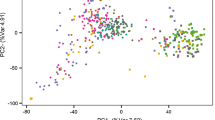
Supplementary Fig. S1. Principal coordinate analysis based on pairwise Rogers’ distances among the 2,325 winter wheat inbred lines. Percentages in brackets indicate the variance contribution of corresponding principal coordinates. (JPEG 6220 kb)
122_2015_2655_MOESM2_ESM.jpg
Supplementary Fig. S2. Distribution of the Rogers’ distances among all pairs of the 2,325 wheat inbred lines. (JPEG 1141 kb)
122_2015_2655_MOESM3_ESM.jpg
Supplementary Fig. S3. Trace plots of residual variance of genomic models based on Bayesian approach (EGBLUP, RKHS and BayesCπ). (JPEG 5045 kb)
122_2015_2655_MOESM4_ESM.jpg
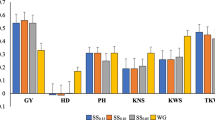
Supplementary Fig. S4. Relationship between the selection accuracy (square root of the broad-sense heritability, h) for grain yield and the number of environments evaluated (combining 2012 and 2013 years data).(JPEG 874 kb)
122_2015_2655_MOESM5_ESM.jpg
Supplementary Fig. S5. Distribution of Best Linear Unbiased Estimates (BLUEs Mg ha−1) of the 2,325 wheat lines evaluated for grain yield as well as linear regression of grain yield on the average number of test environments.(JPEG 1142 kb)
Rights and permissions
About this article
Cite this article
He, S., Schulthess, A.W., Mirdita, V. et al. Genomic selection in a commercial winter wheat population. Theor Appl Genet 129, 641–651 (2016). https://doi.org/10.1007/s00122-015-2655-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-015-2655-1