Abstract
Key message
Yr15 provides broad resistance to stripe rust, an important wheat disease. REMAP- and IRAP-derived co-dominant SCAR markers were developed and localize Yr15 to a 1.2 cM interval. They are reliable across many cultivars.
Abstract
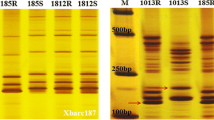
Stripe rust [Pucinia striiformis f.sp. tritici (Pst)] is one of the most important fungal diseases of wheat, found on all continents and in over 60 countries. Wild emmer wheat (Triticum dicoccoides), which is the tetraploid progenitor of durum wheat, is a valuable source of novel stripe rust resistance genes for wheat breeding. T. dicoccoides accession G25 carries Yr15 on chromosome 1BS. Yr15 confers resistance to virtually all tested Pst isolates; it is effective in durum and bread wheat introgressions and their derivatives. Retrotransposons generate polymorphic insertions, which can be scored as Mendelian markers using techniques such as REMAP and IRAP. Six REMAP- and IRAP-derived SCAR markers were mapped using 1,256 F2 plants derived from crosses of the susceptible T. durum accession D447 (DW1) with its resistant BC3F9 and BC3F10 (B9 and B10) near isogenic lines, which carried Yr15 introgressed from G25. The nearest markers segregated 0.1 cM proximally and 1.1 cM distally to Yr15. These markers were also mapped and validated at the same position in another 500 independent F2 plants derived from crosses of B9 and B10 with the susceptible cultivar Langdon (LDN). SC2700 and SC790, defining Yr15 on an interval of 1.2 cM, were found to be reliable and robust co-dominant markers in a wide range of wheat lines and cultivars with and without Yr15. These markers are useful tags in marker-assisted wheat breeding programs that aim to incorporate Yr15 into elite wheat lines and cultivars for durable and broad-spectrum resistance to stripe rust.




Similar content being viewed by others
References
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, Albright LM, Coen DM, Varki A (1995) Current protocols in molecular biology. Wiley, New York
Ballvora A, Jocker A, Viehover P, Ishihara H, Paal J, Meksem K, Bruggmann R, Schoof H, Weisshaar B, Gebhardt C (2007) Comparative sequence analysis of Solanum and Arabidopsis in a hot spot for pathogen resistance on potato chromosome V reveals a patchwork of conserved and rapidly evolving genome segments. BMC Genom 8:112
Breen J, Wicker T, Shatalina M, Frenkel Z, Bertin I, Philippe R, Spielmeyer W, Simkova H, Safar J, Cattonaro F, Scalabrin S, Magni F, Vautrin S, Berges H, International Wheat Genome Sequencing Consortium, Paux E, Fahima T, Dolezel J, Korol A, Feuillet C, Keller. B2013A physical map of the short arm of wheat chromosome 1A. PLoS One 8:e80272
Chague V, Fahima T, Dahan A, Sun GL, Korol AB, Ronin YL, Grama A, Roder MS, Nevo E (1999) Isolation of microsatellite and RAPD markers flanking the Yr15 gene of wheat using NILs and bulked segregant analysis. Genome 42:1050–1056
Chen MX (2005) Epidemiology and control of stripe rust (Puuccinia striformis f.sp. tritici) on wheat. Can J Plant Pathol 27:314–337
Chen XM, Penman L, Wan A, Cheng P (2010) Virulence races of Puccinia striiformis f. sp. tritici in 2006 and 2007 and development of wheat stripe rust and distributions, dynamics, and evolutionary relationships of races from 2000 to 2007 in the United States. Can J Plant Pathol 32:315–333
Delaney DE, Nasuda S, Endo TR, Gill BS, Hulbert SH (1995) Cytologically based physical maps of the group-2 chromosomes of wheat. Theor Appl Genet 91:568–573
Fahima T, Röder M, Grama A, Nevo E (1998) Microsatellite DNA polymorphism divergence in Triticum dicoccoides accessions highly resistant to yellow rust. Theor Appl Genet 96:187–195
FAOSTAT (2012) Production statistics for crops, 2012 data. http://faostat.fao.org/site/567/DesktopDefault.aspx?PageID=567#ancor Accessed 21 Feb 2014
Feuillet C, Travella S, Stein N, Albar L, Nublat A, Keller B (2003) Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc Natl Acad Sci USA 100:15253–15258
Gerechter-Amitai ZK, Grama A (1974) Inheritance of resistance to stripe rust (Puccinia striiformis) in crosses between wild emmer (Triticum dicoccoides) and cultivated tetraploid and hexaploid wheat. I. Triticum durum. Euphytica 23:387–392
Gerechter-Amitai ZK, Stubbs RW (1970) A valuable source of yellow rust resistance in Israeli populations of wild emmer, Triticum dicoccoides Koren. Euphytica 19:12–21
Gerechter-Amitai ZK, Van Silfhout CH, Grama A, Kleitman F (1989a) Yr15: a new gene for resistance to Puccinia striiformis in Triticum dicoccoides sel. G-25. Euphytica 43:187–190
Gerechter-Amitai ZK, Grama A, Van Silfhout CH, Kleitman F (1989b) Resistance to yellow rust in Triticum dicoccoides. II. Crosses with resistant dicoccoides sel. G25. Neth J Plant Pathol 95:79–83
Gill KS, Gill BS, Endo TR, Taylor T (1996) Identification and high-density mapping of gene-rich regions in chromosome group 1 of wheat. Genetics 144:1883–1891
He L, Dooner HK (2009) Haplotype structure strongly affects recombination in a maize genetic interval polymorphic for Helitron and retrotransposon insertions. Proc Natl Acad Sci USA 106:8410–8416
Huang L, Brooks SA, Li W, Fellers JP, Trick HN, Gill BS (2003) Map-based cloning of leaf rust resistance gene Lr21 from the large and polyploid genome of bread wheat. Genetics 164:655–664
Innes RW, Ameline-Torregrosa C, Ashfield T, Cannon E, Cannon SB et al (2008) Differential accumulation of retroelements and diversification of NB-LRR disease resistance genes in duplicated regions following polyploidy in the ancestor of soybean. Plant Physiol 148:1740–1759
International Barley Genome Sequencing Consortium, Mayer KF, Waugh R, Brown JW, Schulman A et al (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491:711–716
Islam MR, Shepherd KW, Mayo GME (1989) Recombination among genes at the L group in flax conferring resistance to rust. Theor Appl Genet 77:540–546
Joshi RK, Nayak S (2013) Perspectives of genomic diversification and molecular recombination towards R-gene evolution in plants. Physiol Mol Biol Plants 19:1–9
Kalendar R, Schulman AH (2006) IRAP and REMAP for retrotransposon-based genotyping and fingerprinting. Nat Protoc 1:2478–2484
Kalendar R, Lee D, Schulman AH (2014) FastPCR software for PCR, in silico PCR, and oligonucleotide assembly and analysis. Methods Mol Biol 1116:271–302
Kota R, Spielmeyer W, McIntosh RA, Lagudah ES (2006) Fine genetic mapping fails to dissociate durable stem rust resistance gene Sr2 from pseudo-black chaff in common wheat (Triticum aestivum L.). Theor Appl Genet 112:492–499
Kronmiller BA, Wise RP (2007) TE nest: Automated chronological annotation and visualization of nested plant transposable elements. Plant Physiol 146:45–59
Laidò G, Mangini G, Taranto F, Gadaleta A, Blanco A, Cattivelli L, Marone D, Mastrangelo AM, Papa R, De Vita P (2013) Genetic diversity and population structure of tetraploid wheats (Triticum turgidum L.) estimated by SSR, DArT and pedigree data. PLoS ONE 8:e67280
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Leigh F, Kalendar R, Lea W, Lee D, Donini P, Schulman AH (2003) Comparison of the utility of barley retrotransposon families for genetic analysis by molecular marker techniques. Mol Genet Genomics 269:464–474
Leister D (2004) Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet 20:116–122
Ling HQ, Zhu Y, Keller B (2003) High-resolution mapping of the leaf rust disease resistance gene Lr1 in wheat and characterization of BAC clones from the Lr1 locus. Theor Appl Genet 106:875–882
Mago R, Miah H, Lawrence GJ, Wellings CR, Spielmeyer W, Bariana HS, McIntosh RA, Pryor AJ, Ellis JG (2005) High-resolution mapping and mutation analysis separate the rust resistance genes Sr31, Lr26 and Yr9 on the short arm of rye chromosome 1. Theor Appl Genet 112:41–50
Manninen O, Kalendar R, Robinson J, Schulman AH (2000) Application of BARE-1 retrotransposon markers to the mapping of a major resistance gene for net blotch in barley. Mol Gen Genet 264:325–334
McIntosh RA, Wellings CR, Park RF (1995) Wheat rusts: an atlas of resistance genes. CSIRO, Melbourne
McIntosh RA, Silk J, The TT (1996) Cytogenetic studies in wheat. XVII. Monosomic analysis and linkage relationships of gene Yr15 for resistance to stripe rust. Euphytica 89:395–399
McIntosh RA, Hart GE, Devos KM, Gale MD, Rogers WJ (1998) Catalogue of gene symbols for wheat. In: Slinkard AE (ed) Proceedings of the 9th international wheat genetics symposium, vol 5. University Extension Press, University of Saskatchewan, Saskatoon, Saskatchewan, Canada, pp 1–235
Michelmore RW, Meyers BC (1998) Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res 8:1113–1130
Murphy LR, Santra D, Kidwell K, Yan G, Chen X, Campbell KG (2009) Linkage maps of wheat stripe rust resistance genes Yr5 and Yr15 for use in marker-assisted selection. Crop Sci 49:1786–1790
Nevo E, Korol AB, Beiles A, Fahima T (2002) Evolution of wild emmer and wheat improvement. Springer, Heidelberg, pp 1–364
Niu YC, Qiao Q, Wu LR (2000) Postulation of resistance genes to stripe rust in commercial wheat cultivars from Henan, Shandong and Anhui provinces. Acta Phytopathol Sin 30:122–128
Paux E, Roger D, Badaeva E, Gay G, Bernard M, Sourdille P, Feuillet C (2006) Characterizing the composition and evolution of homoeologous genomes in hexaploid wheat through BAC-end sequencing on chromosome 3B. Plant J 48:463–474
Peng JH, Fahima T, Röder MS, Huang QY, Dahan A, Li YC, Grama A, Nevo E (2000) High-density molecular map of chromosome region harboring stripe-rust resistance genes YrH52 and Yr15 derived from wild emmer wheat, Triticum dicoccoides. Genetica 109:199–210
Queen RA, Gribbon BM, James C, Jack P, Falvell AJ (2004) Retrotransposon-based molecular markers for linkage and genetic diversity analysis in wheat. Mol Genet Genomics 271:91–97
Ramakrishna W, Emberton J, Ogden M, SanMiguel P, Bennetzen JL (2002) Structural analysis of the maize Rp1 complex reveals numerous sites and unexpected mechanisms of local rearrangement. Plant Cell 14:3213–3223
Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Saintenac C, Faure S, Remay A, Choulet F, Ravel C, Paux E, Balfourier F, Feuillet C, Sourdille P (2011) Variation in crossover rates across a 3-Mb contig of bread wheat (Triticum aestivum) reveals the presence of a meiotic recombination hotspot. Chromosoma 120:185–198
Sandhu D, Champoux JA, Bondareva SN, Gill KS (2001) Identification and physical localization of useful genes and markers to a major gene-rich region on wheat group 1S chromosomes. Genetics 157:1735–1747
Schulman AH, Flavell AJ, Ellis THN, Paux E (2012) The application of LTR retrotransposons as molecular markers in plants. Methods Mol Biol 859:115–153
Shirasu K, Schulman AH, Lahaye T, Schulze-Lefert P (2000) A contiguous 66-kb barley DNA sequence provides evidence for reversible genome expansion. Genome Res 10:908–915
Song WY, Pi LY, Bureau TE, Ronald PC (1998) Identification and characterization of 14 transposon-like elements in the noncoding regions of members of the Xa21 family of disease resistance genes in rice. Mol Gen Genet 258:449–456
Song QJ, Fickus EW, Cregan PB (2002) Characterization of trinucleotide SSR motifs in wheat. Theor Appl Genet 104:286–293
Stein N, Feuillet C, Wicker T, Schlagenhauf E, Keller B (2000) Subgenome chromosome walking in wheat: A 450 kb physical contig in Triticum monococcum L. spans the Lr10 resistance locus in hexaploid wheat (Triticum aestivum L.). Proc Natl Acad Sci USA 97:13436–13441
Sun GL, Fahima T, Korol AB, Turpeinen T, Grama A, Ronin YL, Nevo E (1997) Identification of molecular markers linked to the Yr15 stripe rust resistance gene of wheat originated in wild emmer wheat, Triticum dicoccoides. Theor Appl Genet 95:622–628
Tanhuanpää P, Kalendar R, Schulman AH, Kiviharju E (2007) A major gene for grain cadmium accumulation in oat (Avena sativa L.). Genome 50:588–594
The International Wheat Genome Sequencing Consortium (IWGSC) (2013) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:1251788
Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J (2006) A NAC Gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314:1298–1301
Waugh R, McLean K, Flavell AJ, Pearce SR, Kumar A, Thomas BBT, Powell W (1997) Genetic distribution of Bare-1-like retrotransposable elements in the barley genome revealed by sequence-specific amplification polymorphisms (S-sSAP). Mol Gen Genet 253:687–694
Wei F, Wing RA, Wise RP (2002) Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell 14:1903–1917
White SE, Habera LF, Wesller SR (1994) Retrotransposons in the flanking regions of normal plant genes: A role for copia-like elements in the evolution of gene structure and expression. Proc Natl Acad Sci USA 91:11792–11796
Yan GP, Chen XM, Line RF, Wellings CR (2003) Resistance gene-analog polymorphism markers co-segregating with the Yr5 gene for resistance to wheat stripe rust. Theor Appl Genet 106:636–643
Yaniv E, Raats D, Ronin Y, Korol A, Schulman AH, Fahima T (2014) Comparison of marker-assisted and phenotypic selection for the stripe rust resistance gene Yr15, introgressed from wild emmer wheat. Mol Breed
Yu G-X, Wise RP (2000) An anchored AFLP- and retrotransposon-based map of diploid Avena. Genome 43:736–749
Acknowledgments
This work was supported by the European union’s sixth framework programme (FP6) in the BioExploit project (No. CT-2005-513949). We thank Dr. T. Fahima for his support of, and multiple contributions to, this project. A.-M. Narvanto and U. Lönnqvist are deeply appreciated for technical assistance. For providing plant materials and pedigrees, the authors acknowledge Drs. A. Grama, Z. Gerechter-Amitai, J. Dubcovsky, R. McIntosh, T. The, M.S. Hovmøller. F. Borum, J. Barrett, and B. Schuiling. We thank Dr. J. Manisterski for providing urediniospores of Puccinia striiformis race 38E134 isolate 5006.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The experiments comply with the current laws of the countries in which they were performed.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Mingliang Xu.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Abdollahi Mandoulakani, B., Yaniv, E., Kalendar, R. et al. Development of IRAP- and REMAP-derived SCAR markers for marker-assisted selection of the stripe rust resistance gene Yr15 derived from wild emmer wheat. Theor Appl Genet 128, 211–219 (2015). https://doi.org/10.1007/s00122-014-2422-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-014-2422-8