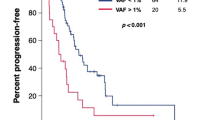
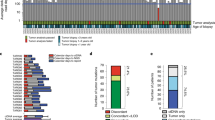
Abstract
Circulating tumor DNA (ctDNA) is considered an ideal sample type for genotyping patients with advanced unresectable cancer to inform treatment decision. It may better capture tumor heterogeneity, especially in gastric adenocarcinoma (GAC). However, there exists little evidence regarding genomic profiling of Chinese advanced GAC patients from ctDNA. Blood samples were obtained from 200 advanced GAC patients. Next-generation sequencing (NGS) was performed on ctDNA using a validated 150-gene panel. Blood tumor mutation burden (bTMB) was calculated according to the NGS results. Blood microsatellite instability (bMSI) status was determined by targeted sequencing of 100 microsatellite loci. One hundred sixty-nine (84.5%) patients carried at least one genomic alteration and 138 (69%) patients had at least one deleterious or likely deleterious alteration (del-alteration). The clonal fraction of del-alterations was higher than that of non-del-alterations (80.1% vs 54.5%, P < 0.0001). The most frequently altered genes were TP53 (38%), LRP1B (20%), MYC (13.5%), ERBB2 (12.5%), and KRAS (11.5%). The alterations were most enriched in the TP53/cell cycle (52%) and the RTK-Ras-MAPK pathway (51.5%). The median bTMB was two (range 0 to 42). Eight patients were identified to be high bMSI, with higher median bTMB than the blood microsatellite stable (bMSS) patients (15 vs 2, P = 0.0062). Patients harboring del-alterations of the DDR pathway had significantly higher percentages of high bTMB and bMSI-H patients than the wild-type subgroup (61.1% vs 6.5%, P < 0.0001; 33.3% vs 1.7%, P = 0.0002). A total of 45.5% cases harbored at least one potentially actionable alteration and one patient achieved complete response after receiving matched targeted therapy. Our study uncovered the molecular characterization of Chinese patients with advanced GAC from ctDNA, including genomic alteration, bTMB, and bMSI status. The findings suggested that targeted NGS-based ctDNA analysis may help inform the clinical decision in advanced GAC.
Key Messages
-
We report the molecular profiling of the largest Chinese advance stage GACs cohort using a CLIA-certified ctDNA assay.
-
Potentially actionable genomic alterations were identified in 45.5% of patients, suggesting clinical utility for ctDNA NGS in advance stage GACs.
-
There was evidence of clinical benefit in one GAC patient with MET amplification treated with MET inhibitor.





Similar content being viewed by others
Availability of data and materials
All data and materials supporting the conclusions of this article were available in the figures, tables, and supplementary materials, which are available to authorized users.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424
Cancer Genome Atlas Research N (2014) Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513(7517):202–209
Wong SS, Kim KM, Ting JC, Yu K, Fu J, Liu S, Cristescu R, Nebozhyn M, Gong L, Yue YG, Wang J, Ronghua C, Loboda A, Hardwick J, Liu X, Dai H, Jin JG, Ye XS, Kang SY, Do IG, Park JO, Sohn TS, Reinhard C, Lee J, Kim S, Aggarwal A (2014) Genomic landscape and genetic heterogeneity in gastric adenocarcinoma revealed by whole-genome sequencing. Nat Commun 5:5477
Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, Ye XS, Do IG, Liu S, Gong L, Fu J, Jin JG, Choi MG, Sohn TS, Lee JH, Bae JM, Kim ST, Park SH, Sohn I, Jung SH, Tan P, Chen R, Hardwick J, Kang WK, Ayers M, Hongyue D, Reinhard C, Loboda A, Kim S, Aggarwal A (2015) Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 21(5):449–456
Kim ST, Cristescu R, Bass AJ, Kim KM, Odegaard JI, Kim K, Liu XQ, Sher X, Jung H, Lee M, Lee S, Park SH, Park JO, Park YS, Lim HY, Lee H, Choi M, Talasaz A, Kang PS, Cheng J, Loboda A, Lee J, Kang WK (2018) Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med 24(9):1449–1458
Nagaraja AK, Kikuchi O, Bass AJ (2019) Genomics and targeted therapies in gastroesophageal adenocarcinoma. Cancer Discov 9(12):1656–1672
Janjigian YY, Sanchez-Vega F, Jonsson P, Chatila WK, Hechtman JF, Ku GY, Riches JC, Tuvy Y, Kundra R, Bouvier N, Vakiani E, Gao J, Heins ZJ, Gross BE, Kelsen DP, Zhang L, Strong VE, Schattner M, Gerdes H, Coit DG, Bains M, Stadler ZK, Rusch VW, Jones DR, Molena D, Shia J, Robson ME, Capanu M, Middha S, Zehir A, Hyman DM, Scaltriti M, Ladanyi M, Rosen N, Ilson DH, Berger MF, Tang L, Taylor BS, Solit DB, Schultz N (2018) Genetic predictors of response to systemic therapy in esophagogastric cancer. Cancer Discov 8(1):49–58
Parikh AR, Leshchiner I, Elagina L, Goyal L, Levovitz C, Siravegna G, Livitz D, Rhrissorrakrai K, Martin EE, Van Seventer EE, Hanna M, Slowik K, Utro F, Pinto CJ, Wong A, Danysh BP, de la Cruz FF, Fetter IJ, Nadres B, Shahzade HA, Allen JN, Blaszkowsky LS, Clark JW, Giantonio B, Murphy JE, Nipp RD, Roeland E, Ryan DP, Weekes CD, Kwak EL, Faris JE, Wo JY, Aguet F, Dey-Guha I, Hazar-Rethinam M, Dias-Santagata D, Ting DT, Zhu AX, Hong TS, Golub TR, Iafrate AJ, Adalsteinsson VA, Bardelli A, Parida L, Juric D, Getz G, Corcoran RB (2019) Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat Med 25(9):1415–1421
Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R, Rosenfeld N (2017) Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 17(4):223–238
Heitzer E, Haque IS, Roberts CES, Speicher MR (2019) Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet 20(2):71–88
Administration TUSFaD (2020) FDA approves Foundation Medicine’s FoundationOne®Liquid CDx, a comprehensive pan-tumor liquid biopsy test with multiple companion diagnostic indications for patients with advanced cancer. https://www.foundationmedicine.com/press-releases/445c1f9e-6cbb-488b-84ad-5f133612b721. Accessed Accessed 26 Aug 2020
Administration TUSFaD (2020) FDA approves first liquid biopsy next-generation sequencing companion diagnostic test. https://www.fda.gov/news-events/press-announcements/fda-approves-first-liquid-biopsy-next-generation-sequencing-companion-diagnostic-test. Accessed Accessed 7 Aug 2020
Wright JR Jr (2013) Charles Emmanuel Sedillot and Emile Kuss: the first cancer biopsy. Int J Surg 11(1):106–107
Sawada K, Kotani D, Bando H (2018) The clinical landscape of circulating tumor DNA in gastrointestinal malignancies. Front Oncol 8:263
Wang DS, Liu ZX, Lu YX, Bao H, Wu X, Zeng ZL, Liu Z, Zhao Q, He CY, Lu JH, Wang ZQ, Qiu MZ, Wang F, Wang FH, Li YH, Wang XN, Xie D, Jia WH, Shao YW, Xu RH (2019) Liquid biopsies to track trastuzumab resistance in metastatic HER2-positive gastric cancer. Gut 68(7):1152–1161
Kato S, Okamura R, Baumgartner JM, Patel H, Leichman L, Kelly K, Sicklick JK, Fanta PT, Lippman SM, Kurzrock R (2018) Analysis of circulating tumor DNA and clinical correlates in patients with esophageal, gastroesophageal junction, and gastric adenocarcinoma. Clin Cancer Res 24(24):6248–6256
Cabel L, Proudhon C, Romano E, Girard N, Lantz O, Stern MH, Pierga JY, Bidard FC (2018) Clinical potential of circulating tumour DNA in patients receiving anticancer immunotherapy. Nat Rev Clin Oncol 15(10):639–650
Sabari JK, Offin M, Stephens D, Ni A, Lee A, Pavlakis N, Clarke S, Diakos CI, Datta S, Tandon N, Martinez A, Myers ML, Makhnin A, Leger Y, Yu HA, Paik PK, Chaft JE, Kris MG, Jeon JO, Borsu LA, Ladanyi M, Arcila ME, Hernandez J, Henderson S, Shaffer T, Garg K, DiPasquo D, Raymond CK, Lim LP, Li M, Hellmann MD, Drilon A, Riely GJ, Rusch VW, Jones DR, Rimner A, Rudin CM, Isbell JM, Li BT (2018) A prospective study of circulating tumor DNA to guide matched targeted therapy in lung cancers. J Natl Cancer Inst 111:575–583
Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, Azad NS, Bardelli A, Brem H, Cameron JL, Lee CC, Fecher LA, Gallia GL, Gibbs P, Le D, Giuntoli RL, Goggins M, Hogarty MD, Holdhoff M, Hong SM, Jiao Y, Juhl HH, Kim JJ, Siravegna G, Laheru DA, Lauricella C, Lim M, Lipson EJ, Marie SK, Netto GJ, Oliner KS, Olivi A, Olsson L, Riggins GJ, Sartore-Bianchi A, Schmidt K, Shih LM, Oba-Shinjo SM, Siena S, Theodorescu D, Tie J, Harkins TT, Veronese S, Wang TL, Weingart JD, Wolfgang CL, Wood LD, Xing D, Hruban RH, Wu J, Allen PJ, Schmidt CM, Choti MA, Velculescu VE, Kinzler KW, Vogelstein B, Papadopoulos N, Diaz LA Jr (2014) Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 6(224):224ra224
Maron SB, Chase LM, Lomnicki S, Kochanny S, Moore KL, Joshi S, Landron S, Johnson J, Kiedrowski LA, Nagy RJ, Lanman RB, Kim ST, Lee J, Catenacci DVT (2019) Circulating tumor DNA sequencing analysis of gastroesophageal adenocarcinoma. Clin Cancer Res 25:7098–7112
Schrock AB, Pavlick D, Klempner SJ, Chung JH, Forcier B, Welsh A, Young L, Leyland-Jones B, Bordoni R, Carvajal RD, Chao J, Kurzrock R, Sicklick JK, Ross JS, Stephens PJ, Devoe C, Braiteh F, Ali SM, Miller VA (2018) Hybrid capture–based genomic profiling of circulating tumor DNA from patients with advanced cancers of the gastrointestinal tract or anus. Clin Cancer Res 24(8):1881–1890
Koldby KM, Mortensen MB, Detlefsen S, Pfeiffer P, Thomassen M, Kruse TA (2019) Tumor-specific genetic aberrations in cell-free DNA of gastroesophageal cancer patients. J Gastroenterol 54(2):108–121
Wang Z, Duan J, Cai S, Han M, Dong H, Zhao J, Zhu B, Wang S, Zhuo M, Sun J, Wang Q, Bai H, Han J, Tian Y, Lu J, Xu T, Zhao X, Wang G, Cao X, Li F, Wang D, Chen Y, Bai Y, Zhao J, Zhao Z, Zhang Y, Xiong L, He J, Gao S, Wang J (2019) Assessment of blood tumor mutational burden as a potential biomarker for immunotherapy in patients with non-small cell lung cancer with use of a next-generation sequencing cancer gene panel. JAMA Oncol 5(5):696–702
Wang Z, Qin H, Wang M, Gong J, Wang X, Li J, Gao J, Li Z, Wang D, Cai S, Bai Y, Xiong L, Li F, Shen L (2018) bMSI better predicts the responses to immune checkpoint inhibitors (ICI) than MMR/MSI from historical tissue specimens in metastatic gastrointestinal cancer patients. Ann Oncol 29:viii28
Strickler JH, Loree JM, Ahronian LG, Parikh AR, Niedzwiecki D, Pereira AAL, McKinney M, Korn WM, Atreya CE, Banks KC, Nagy RJ, Meric-Bernstam F, Lanman RB, Talasaz A, Tsigelny IF, Corcoran RB, Kopetz S (2018) Genomic landscape of cell-free DNA in patients with colorectal cancer. Cancer Discov 8(2):164–173
Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, Rudolph JE, Yaeger R, Soumerai T, Nissan MH, Chang MT, Chandarlapaty S, Traina TA, Paik PK, Ho AL, Hantash FM, Grupe A, Baxi SS, Callahan MK, Snyder A, Chi P, Danila D, Gounder M, Harding JJ, Hellmann MD, Iyer G, Janjigian Y, Kaley T, Levine DA, Lowery M, Omuro A, Postow MA, Rathkopf D, Shoushtari AN, Shukla N, Voss M, Paraiso E, Zehir A, Berger MF, Taylor BS, Saltz LB, Riely GJ, Ladanyi M, Hyman DM, Baselga J, Sabbatini P, Solit DB, Schultz N (2017) OncoKB: a precision oncology knowledge base. JCO Precis Oncol 2017:1–16
Chen K, Yang D, Li X, Sun B, Song F, Cao W, Brat DJ, Gao Z, Li H, Liang H, Zhao Y, Zheng H, Li M, Buckner J, Patterson SD, Ye X, Reinhard C, Bhathena A, Joshi D, Mischel PS, Croce CM, Wang YM, Raghavakaimal S, Li H, Lu X, Pan Y, Chang H, Ba S, Luo L, Cavenee WK, Zhang W, Hao X (2015) Mutational landscape of gastric adenocarcinoma in Chinese: implications for prognosis and therapy. Proc Natl Acad Sci U S A 112(4):1107–1112
Xing R, Zhou Y, Yu J, Yu Y, Nie Y, Luo W, Yang C, Xiong T, Wu WKK, Li Z, Bing Y, Lin S, Zhang Y, Hu Y, Li L, Han L, Yang C, Huang S, Huang S, Zhou R, Li J, Wu K, Fan D, Tang G, Dou J, Zhu Z, Ji J, Fang X, Lu Y (2019) Whole-genome sequencing reveals novel tandem-duplication hotspots and a prognostic mutational signature in gastric cancer. Nat Commun 10(1):2037
Seo HM, Chang YS, Joo SH, Kim YW, Park YK, Hong SW, Lee SH (2009) Clinicopathologic characteristics and outcomes of gastric cancers with the MSI-H phenotype. J Surg Oncol 99(3):143–147
Jung JH, Lee H, Cao B, Liao P, Zeng SX, Lu H (2019) RNA-binding motif protein 10 induces apoptosis and suppresses proliferation by activating p53. Oncogene. 39:1031–1040
Hernandez J, Bechara E, Schlesinger D, Delgado J, Serrano L, Valcarcel J (2016) Tumor suppressor properties of the splicing regulatory factor RBM10. RNA Biol 13(4):466–472
Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J, Mollaee M, Wagner KU, Koduru P, Yopp A, Choti MA, Yeo CJ, McCue P, White MA, Knudsen ES (2015) Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 6:6744
Song Y, Li L, Ou Y, Gao Z, Li E, Li X, Zhang W, Wang J, Xu L, Zhou Y, Ma X, Liu L, Zhao Z, Huang X, Fan J, Dong L, Chen G, Ma L, Yang J, Chen L, He M, Li M, Zhuang X, Huang K, Qiu K, Yin G, Guo G, Feng Q, Chen P, Wu Z, Wu J, Ma L, Zhao J, Luo L, Fu M, Xu B, Chen B, Li Y, Tong T, Wang M, Liu Z, Lin D, Zhang X, Yang H, Wang J, Zhan Q (2014) Identification of genomic alterations in oesophageal squamous cell cancer. Nature 509(7498):91–95
Zhou C, Ye M, Ni S, Li Q, Ye D, Li J, Shen Z, Deng H (2018) DNA methylation biomarkers for head and neck squamous cell carcinoma. Epigenetics 13(4):398–409
Suehara Y, Arcila M, Wang L, Hasanovic A, Ang D, Ito T, Kimura Y, Drilon A, Guha U, Rusch V, Kris MG, Zakowski MF, Rizvi N, Khanin R, Ladanyi M (2012) Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin Cancer Res 18(24):6599–6608
Gu TL, Deng X, Huang F, Tucker M, Crosby K, Rimkunas V, Wang Y, Deng G, Zhu L, Tan Z, Hu Y, Wu C, Nardone J, MacNeill J, Ren J, Reeves C, Innocenti G, Norris B, Yuan J, Yu J, Haack H, Shen B, Peng C, Li H, Zhou X, Liu X, Rush J, Comb MJ (2011) Survey of tyrosine kinase signaling reveals ROS kinase fusions in human cholangiocarcinoma. PLoS One 6(1):e15640. https://doi.org/10.1371/journal.pone.0015640
Hirotsu Y, Nakagomi H, Amemiya K, Oyama T, Inoue M, Mochizuki H, Omata M (2017) Intrinsic HER2 V777L mutation mediates resistance to trastuzumab in a breast cancer patient. Med Oncol 34(1):3
Kim ST, Banks KC, Pectasides E, Kim SY, Kim K, Lanman RB, Talasaz A, An J, Choi MG, Lee JH, Sohn TS, Bae JM, Kim S, Park SH, Park JO, Park YS, Lim HY, Kim NKD, Park W, Lee H, Bass AJ, Kim K, Kang WK, Lee J (2018) Impact of genomic alterations on lapatinib treatment outcome and cell-free genomic landscape during HER2 therapy in HER2+ gastric cancer patients. Ann Oncol 29(4):1037–1048
Eroglu Z, Zaretsky JM, Hu-Lieskovan S, Kim DW, Algazi A, Johnson DB, Liniker E, Ben K, Munhoz R, Rapisuwon S, Gherardini PF, Chmielowski B, Wang X, Shintaku IP, Wei C, Sosman JA, Joseph RW, Postow MA, Carlino MS, Hwu WJ, Scolyer RA, Messina J, Cochran AJ, Long GV, Ribas A (2018) High response rate to PD-1 blockade in desmoplastic melanomas. Nature 553(7688):347–350
Hellmann MD, Ciuleanu TE, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C, Minenza E, Linardou H, Burgers S, Salman P, Borghaei H, Ramalingam SS, Brahmer J, Reck M, O’Byrne KJ, Geese WJ, Green G, Chang H, Szustakowski J, Bhagavatheeswaran P, Healey D, Fu Y, Nathan F, Paz-Ares L (2018) Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med 378(22):2093–2104
Wang F, Wei XL, Wang FH, Xu N, Shen L, Dai GH, Yuan XL, Chen Y, Yang SJ, Shi JH, Hu XC, Lin XY, Zhang QY, Feng JF, Ba Y, Liu YP, Li W, Shu YQ, Jiang Y, Li Q, Wang JW, Wu H, Feng H, Yao S, Xu RH (2019) Safety, efficacy and tumor mutational burden as a biomarker of overall survival benefit in chemo-refractory gastric cancer treated with toripalimab, a PD-1 antibody in phase Ib/II clinical trial NCT02915432. Ann Oncol 30(9):1479–1486
Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, Wong F, Azad NS, Rucki AA, Laheru D, Donehower R, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Greten TF, Duffy AG, Ciombor KK, Eyring AD, Lam BH, Joe A, Kang SP, Holdhoff M, Danilova L, Cope L, Meyer C, Zhou S, Goldberg RM, Armstrong DK, Bever KM, Fader AN, Taube J, Housseau F, Spetzler D, Xiao N, Pardoll DM, Papadopoulos N, Kinzler KW, Eshleman JR, Vogelstein B, Anders RA, Diaz LA Jr (2017) Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357(6349):409–413
Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, Biedrzycki B, Donehower RC, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Duffy SM, Goldberg RM, de la Chapelle A, Koshiji M, Bhaijee F, Huebner T, Hruban RH, Wood LD, Cuka N, Pardoll DM, Papadopoulos N, Kinzler KW, Zhou S, Cornish TC, Taube JM, Anders RA, Eshleman JR, Vogelstein B, Diaz LA Jr (2015) PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 372(26):2509–2520
Gandara DR, Paul SM, Kowanetz M, Schleifman E, Zou W, Li Y, Rittmeyer A, Fehrenbacher L, Otto G, Malboeuf C, Lieber DS, Lipson D, Silterra J, Amler L, Riehl T, Cummings CA, Hegde PS, Sandler A, Ballinger M, Fabrizio D, Mok T, Shames DS (2018) Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med 24(9):1441–1448
Willis J, Lefterova MI, Artyomenko A, Kasi PM, Nakamura Y, Mody K, Catenacci DVT, Fakih M, Barbacioru C, Zhao J, Sikora M, Fairclough SR, Lee H, Kim KM, Kim ST, Kim J, Gavino D, Benavides M, Peled N, Nguyen T, Cusnir M, Eskander RN, Azzi G, Yoshino T, Banks KC, Raymond VM, Lanman RB, Chudova DI, Talasaz A, Kopetz S, Lee J, Odegaard JI (2019) Validation of microsatellite instability detection using a comprehensive plasma-based genotyping panel. Clin Cancer Res 25:7035–7045
Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, Schrock A, Campbell B, Shlien A, Chmielecki J, Huang F, He Y, Sun J, Tabori U, Kennedy M, Lieber DS, Roels S, White J, Otto GA, Ross JS, Garraway L, Miller VA, Stephens PJ, Frampton GM (2017) Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 9(1):34
Parikh AR, He Y, Hong TS, Corcoran RB, Clark JW, Ryan DP, Zou L, Ting DT, Catenacci DV, Chao J, Fakih M, Klempner SJ, Ross JS, Frampton GM, Miller VA, Ali SM, Schrock AB (2019) Analysis of DNA damage response gene alterations and tumor mutational burden across 17,486 tubular gastrointestinal carcinomas: implications for therapy. Oncologist 24(10):1340–1347
Tubbs A, Nussenzweig A (2017) Endogenous DNA damage as a source of genomic instability in cancer. Cell 168(4):644–656
Acknowledgements
The authors thank the patients who participate in this study and the members who involved in the data collection and analysis.
Funding
This study was financially supported by the Capital Health Research and Development of Special (grant number 2018-2-1023) and the Pilot Project (3rd Round) to Reform Public Development of Beijing Municipal Medical Research Institutes (grant number 2019-1).
Author information
Authors and Affiliations
Contributions
Meng Zhang, Hui Chen, Xiaochen Zhao, Yuezong Bai, Shuqin Jia, and Jiafu Ji conceived and designed the study. Changsong Qi, Zhenghang Wang, and Hui Chen performed the research. Meng Zhang, Hui Chen, Xueming Zhang, Yifan Zhou, Chan Gao, and Xiaochen Zhao wrote the paper. Changsong Qi and Hui Chen analyzed the data. All authors contributed to manuscript revision, read, and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This study was conducted according to the Declaration of Helsinki and was approved by the Institutional Ethical Standards Committee of Peking University Cancer Hospital and Institute (NO.2016XJS01).
Informed consent
Written informed consent was obtained from each patient.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Figure S1
Comparison of alteration frequencies between 3DMed cohort and MSK-UC cohort (A) and MSK-SMC cohort (B). (PNG 44 kb)
Figure S2
Correlation between SNV/indels number and MSI status. (PNG 17 kb)
Figure S3
Number of deleterious DDR alterations (PNG 22 kb)
Table S1
Panel gene list (XLSX 10 kb)
Table S2
Case presentation of patient with target therapy based on ctDNA (XLSX 9 kb)
Table. S3
anti-ERBB2 resistance related alterations in ERBB2+ GACs (XLSX 9 kb)
Rights and permissions
About this article
Cite this article
Zhang, M., Qi, C., Wang, Z. et al. Molecular characterization of ctDNA from Chinese patients with advanced gastric adenocarcinoma reveals actionable alterations for targeted and immune therapy. J Mol Med 99, 1311–1321 (2021). https://doi.org/10.1007/s00109-021-02093-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-021-02093-z