Abstract
The vernal pool fairy shrimp (Branchinecta lynchi) (Eng et al. in J Crustac Biol 10:247–277, 1990) is broadly distributed throughout California’s Central Valley with disjunct populations in southern California and southern Oregon. A survey of genetic variation at the mitochondrial cytochrome c oxidase subunit I (COI) gene was conducted from individuals collected throughout the range of the species. Phylogenetic analysis of unique haplotypes could not resolve any significant genetic partitions below the species level. A detailed analysis of samples from two pools indicates that sample sizes underestimate overall variation, but that the general phylogeographic pattern still holds. Evidence was found for a putative long-distance dispersal event between Central Valley sites and the Oregon site. These results indicate that geographically limiting stochastic colonization followed by local diversification may be important in governing current genetic structure for this species. Future and current conservation/restoration efforts should recognize the geographic limitations to population structure for this species and focus on local endemic genetic variation.
Similar content being viewed by others
Introduction
Dispersal, historical colonization and past climatic/geologic events are important forces in determining contemporary genetic population structure in natural populations (Avise 2004). In passively dispersed aquatic invertebrates, non-equilibrium processes like long-distance dispersal and stochastic colonization events, often govern patterns of genetic differentiation over regional and continental scales (Boileau et al. 1992; Hebert et al. 2003; Ketmaier et al. 2005; Gomez et al. 2007). Previous work has also found evidence for isolation by distance at local scales (Bohonak 1998; Brendonck et al. 2000; Hulsmans et al. 2007), but it is unknown if these patterns are due to equilibrium processes. Little work has been done on the range-wide genetic structure of passively dispersed aquatic organisms with limited geographic distributions and more work is needed to address how patterns of differentiation for such species can inform conservation efforts.
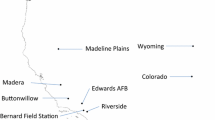
Western North America possesses a high degree of endemism for fairy shrimp of the genus Branchinecta (Anostraca) (Belk and Brtek 1995; Belk and Fugate 2000; Belk and Rogers 2002). Branchinecta fairy shrimp predominately inhabit seasonal wetlands, which are currently being lost in California at an extremely high rate due to development and degradation by human activities (Belk 1998). B. lynchi (Eng et al. 1990) has the widest distribution for any of the CA endemic Branchinecta, found in the Santa Rosa Plateau, Santa Barbara County, Ventura County, the Coast Ranges of Monterey County, throughout the great Central Valley, and in the Agate Desert of southwestern Oregon (Fig. 1). This species was only recently described by Eng et al. (1990). B. lynchi inhabits a variety of vernal pool habitats, from clear, small to moderately sized vernal pools and swales to larger turbid vernal playas (Eriksen and Belk 1999). Current threats to the long-term persistence of B. lynchi are habitat loss and alteration, though it is unknown how shifts in climate regimes due to regional and global climate change may influence species survival. Currently this species is listed as threatened by the US Fish and Wildlife Service (USFWS 1994). To understand the genetic structure of this narrow-range endemic fairy shrimp, I investigated the range-wide phylogeography of B. lynchi. This information will provide a framework for establishing conservation priorities for these imperiled anostrocans.
Map of B. lynchi distribution and sampling locations. Shaded areas represent the known distribution of B. lynchi. Grey is the San Joaquin Valley, black is the Sacramento Valley, horizontal lines (site 1) are southern Oregon, diagonal lines (site 22) are Monterey, and white (site 25) is the Santa Rosa Plateau. Specifics of the site codes are listed in Table 1
Methods
Sampling
The majority of B. lynchi samples were obtained from the California Academy of Sciences (CAS) collection (Table 1). Museum specimens were originally collected from 1998 to 2002. I also sampled individuals from two pools from eastern Merced County in 2009, Pool 6 and Pool 11. These were ‘intensively’ sampled, containing 23 and 25 individuals, respectively. All individuals from CAS were stored in 80–95% ethanol. Whole individuals were rinsed in milliQ water and macerated in 400 μl of 2× CTAB buffer and incubated at 65°C for 1 h. Samples were extracted once with 500 μl of phenol:chloroform:isoamyl alcohol (25:24:1) and once with 500 μl of chloroform:isoamyl alcohol (24:1). DNA was precipitated in 1 ml of ice cold ethanol and 30 μl of 3 M sodium acetate at −20°C overnight. Following centrifugation, DNA pellets were washed with 75% ethanol and air dried. The resulting DNA pellet was re-suspended in 100 μl of milliQ water.
A 439 base pair fragment of the mitochondrial COI gene was amplified with the newly designed primers COIc-F1 (5′-AAT GTT AAT TCG GGC AGA-3′) and COIc-R1 (5′-GAA AGA AGA AGG AGA ACA GC-3′). Each PCR reaction contained 2 μl of 1:10 diluted DNA, 1× Amplitaq PCR buffer (Applied Biosystem Inc.), 2.5 mM MgCl2, 0.1 mM dNTPS, 0.3 pmol of each primer and 0.5 U of Amplitaq (Applied Biosystem Inc.) in a 50 μl reaction. Thermocycling was done on an Applied Biosystems 2720 cycler under the following temperature profile: initial denaturation for 3 min at 94°C, 30 cycles at 94°C for 30 s, 52°C for 30 s and 72°C for 30 s, followed by a 5 min extension at 72°C. PCR products were cleaned with the Qiaquick PCR Purification kit (Qiagen Inc) and sequenced on an ABI3730 automated sequencer. Trace files were imported into Sequencher (GeneCodes, Inc.) and aligned by eye. Four Santa Rosa Plateau sequences were kindly provided by A. Bohonak (San Diego State University, USA Genbank Accession numbers: FJ439749, FJ439750, FJ439753, FJ439752).
To determine how sampling effort influences the number of haplotypes discovered, rarefaction curves were generated for the two intensively sampled pools from eastern Merced County. Rarefaction curves were estimated by determining the cumulative number of haplotypes sampled with increased sampling effort (number of individuals). Curves were estimated with the program ANALYTIC RAREFACTION v1.3 (http://www.uga.edu/strata/software/) and asymptotes of each curve were used to assess to total number of haplotypes expected with increased sampling effort (Kohn et al. 1999).
Nucleotide substitution model evaluation, for the unique haplotypes and outgroup taxa, was done using the program MRMODELTEST v2.3 (Nylander 2004). The GTR model with a proportion of invariant sites (0.745) was the best model indicated by the Aikake Information Criterion (AIC). Phylogenetic relationship of unique haplotypes was determined using Bayesian and maximum likelihood (ML) frameworks. I used the programs MRBAYES v2.3 (Ronquist and Huelsenbeck 2003) and PHYML v2.4.4 (Guindon and Gascuel 2003) to construct Bayesian and ML trees, respectively. For the Bayesian analysis, 10,000,000 generations were run, sampling trees every 1,000 generations. The first 2,500 trees were discarded as burn-in. The remaining trees were used to construct a 50% majority consensus tree. The ML tree was constructed using the AIC model parameters and node support was assessed with 1,000 non-parametric bootstrap replicates. A haplotype network was constructed using the neighbor-net algorithm (Huson and Bryant 2006) in the software package SPLITSTREE v4.11.3 (http://www.splitstree.org). Outgroups in the MRBAYES and PHYML analyses included B. lindahli and B. sandiegonensis sequences that were downloaded from GENBANK (FJ439744 and FJ439689). These species are closely related to B. lynchi based on previous work (Fugate 1992).
Results
A total of 39 COI haplotypes were obtained from 107 B. lynchi individuals (HM568467-HM568505). Some 29 of these haplotypes (A-AC) were isolated from 59 museum typed individuals. The Pool 6 and Pool 11 samples contained seven and three unique haplotypes, respectively. The haplotypes found in Pools 6 and 11 were not shared among any of the museum samples. One of the two ‘intensively’ sampled pools contained a high number of unique haplotypes (Pool 6). Rarefaction curves indicate that the estimate for the number of haplotypes in Pool 6 was underestimated given the number of individuals sampled (asymptote of curve = 9.7; Fig. 2). While the number of haplotypes discovered in pool 11 was near the maximum number expected based on the rarefaction analysis (asymptote of curve = 3.5; Fig. 2).
The Bayesian phylogenetic analysis did not show any strong geographic association of haplotypes (Fig. 3). However, three clades (<0.8 posterior probability for Bayesian analysis and <50% bootstrap support for the ML tree) were recovered. These same clades were recovered in the ML analysis with low bootstrap support, though the overall topology was slightly different (not shown). Clade I contained haplotypes from sites 2, 3, 5, 10, 11, and 18 (southern Sacramento Valley + San Joaquin Valley). Clade II haplotypes were from sites 3, 12–15, 19 and 21 (northern San Joaquin Valley and Sacramento Valley), and Clade III haplotypes were found in sites 5–8 (central San Joaquin Valley). Sequences from Santa Rose Plateau site did not group out as monophyletic (haplotypes BLyn02, 04, 06 and 18). Similar patterns were observed in the neighbor-net network (Fig. 4), with all three clades being recovered and the lack of any strong association of haplotypes from the Santa Rosa Plateau.
Discussion
The lack of a strong phylogeographic relationship of haplotypes is somewhat surprising for this species. In contrast, other phylogeographic studies of anostracans often find deep within species divergences (Ketmaier et al. 2005, 2008; Munoz et al. 2008), although most of these species have much larger geographic ranges than B. lynchi. I observed three clades in the Bayesian, ML and network analyses, but support for these clades was low in both analyses. If we consider the geographic distribution of these clades there is a hint of some phylogeographic signal. Clade I is restricted to the northern San Joaquin and Sacramento Valleys, Clade II is found throughout the Sacramento Valley (with one site in the northern San Joaquin Valley), and Clade III is restricted to the central portion of the San Joaquin Valley. Given the spatial and site overlap for haplotypes from these clades, the observed pattern for B. lynchi indicates that independent historical colonization events, geographically limited dispersal, and sub-regional diversification may be the overriding mechanisms determining phylogeographic patterns in B. lynchi. It also suggests that the central portion of the great Central Valley may have served as a historical site for diversification throughout the San Joaquin and Sacramento Valleys.
I also observed a number of haplotypes that were not phylogenetically affiliated with any of the aforementioned clades that were restricted to specific sites (haplotypes I, P, W, BLyn02, BLyn04, BLyn09 & BLyn18). Even though a weak signature of geographic differentiation was observed, a large number of unrelated haplotypes display regional endemism. Given this scenario, larger genetic units below the species level (evolutionary significant units or management units) will be difficult to define solely on the use of mitochondrial DNA for this species.
The distribution of haplotype ‘A’, which was found in individuals from the Sacramento/San Joaquin Valleys and southern Oregon, suggests that a long-distance colonization event may have occurred between these sites. This phenomenon has been observed in other anostracan species (Ketmaier et al. 2008; Munoz et al. 2008) and may be ubiquitous within the group. The range of this species lies within the western portion of the Pacific Flyway, which would provide ample opportunity for waterfowl or shorebird mediated dispersal. It is still unclear how common these type of dispersal events are and how important they might be in generating species-wide phylogeograhic patterns, especially for narrow range endemics like B. lynchi.
The rarefaction curves suggest that the low sample sizes used for most sites in this study probably greatly underrepresented the ‘true’ haplotypic diversity on a per pool basis. For instance, larger number of haplotypes was discovered in Pool 6. However, this does not undermine the observed phylogeographic patterns in the data set. If in fact more haplotypes are to be discovered with a more thorough sampling (on a per pool basis), one would expect to see a further weakening of the currently observed clades and a higher probability of uncovering haplotypes that are shared among regions. This would suggest that this species has a much higher propensity for movement, in an evolutionary sense, than shown here. Despite limited sampling from the museum collection, a similar result was found when both the museum and the two intensively sampled pools are considered separately: lineages from Clades I and III recovered from mid-Central Valley sites. It still remains to be seen how much intra-pool variation exists, and for other closely related species, and what factors influence the amount of within pool variation.
Aside from the sampling limitations, which don’t appear to influence the overall phylogeographic signal, two additional factors could influence the observed results for this species; i.e. sampling over multiple years and the potential for alternative reproductive modes (parthenogenesis). The latter is especially crucial given the potentially large resting egg bank and bet hedging strategy that most anostracans utilize (Simovich and Hathaway 1997). While I am unable to address these issues in this study, future work should look to delineate temporal aspects of population structure for this and other species with similar life histories. Initial work on this species indicates that haplotype frequencies are stable over short time periods (A. Aguilar, unpubl. data). Regardless, long-term evolutionary relationships across regional scales should not be greatly influenced by sampling over shorter time periods. The presence of parthenogenetic individuals could greatly alter the expectations of phylogeographic structure. Though parthenogenesis has been documented in Artemia, it is not known for any species of Branchinecta (DC Rogers and JL Krumm, personal communication).
These results suggest that conservation and restoration actions for this species should be determined at local scales. Western North America has a number of endemic Branchinecta species that all share a close phylogenetic relationship (B. conservatio, B. longiantenna, B. mesovallensis, and B. sandiegoensis—Fugate 1992; Aguilar and Rogers, unpubl. data) and the results presented here have implications for the sub-specific diversification and conservation for these species. The large amount of regional endemism for mitochondrial haplotypes suggests that conservation and management need to take into account local evolutionary and ecological dynamics. This fact also suggests that other anostracans with similar distributions and ecological characteristics could be similarly managed.
References
Avise JC (2004) Molecular Markers, Natural History and Evolution, 2nd edn. Sinauer, Sunderland
Belk D (1998) Global status and trends in ephemeral pool invertebrate conservation: implications for Californian Fairy Shrimp. In: Witham CW, Bauder ET, Belk D, Ferren WR Jr, Ornduff R (eds) Ecology, conservation, and management of vernal pool ecosystems—Proceedings from a 1996 Conference. California Native Plant Society, Sacramento, pp 147–150
Belk D, Brtek J (1995) Checklist of the Anostraca. Hydrobiologia 298:315–353
Belk D, Fugate M (2000) Two new Branchinecta (Crustacea:Anostraca) from the southwestern United States. Southwest Nat 45:111–117
Belk D, Rogers DC (2002) A confusing trio of Branchinecta (Crustacea:Anostraca) from the western North America with a description of a new species. Hydrobiologia 486:49–55
Bohonak AJ (1998) Genetic population structure of the fairy shrimp Branchinecta coloradensis (Anostraca) in the Rocky Mountains of Colorado. Can J Zool 76:2049–2057
Boileau MG, Hebert PDN, Schwartz SS (1992) Nonequilibrium gene frequency divergence—persistent founder effects in natural populations. J Evol Biol 5:25–39
Brendonck L, De Meester L, Riddoch BJ (2000) Regional structuring of genetic variation in short-lived rock pool populations of Branchipodopsis wolfi (Crustacea:Anostraca). Oecologia 123:506–515
Eng LL, Belk D, Eriksen CH (1990) Californian Anostraca—distribution, habitat, and status. J Crustac Biol 10:247–277
Eriksen C, Belk D (1999) Fairy shrimps of California’s puddles, pools and playas. Mad River Press, Eureka
Fugate ML (1992) Speciation in the fairy shrimp genus Branchinecta (Crustacea:Anostraca) from North America. PhD Dissertation. University of California, Riverside
Gomez A, Montero-Pau J, Lunt DH, Serra M, Campillo S (2007) Persistent genetic signatures of colonization in Brachionus manjavacas rotifers in the Iberian Peninsula. Mol Ecol 16:3228–3240
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Hebert PDN, Witt JDS, Adamowicz SJ (2003) Phylogeographical patterning in Daphnia ambigua: regional divergence and intercontinental cohesion. Limnol Oceanogr 48:261–268
Hulsmans A, Moreau K, De Meester L, Riddoch BJ, Brendonck L (2007) Direct and indirect measures of dispersal in the fairy shrimp Branchipodopsis wolfi indicate a small-scale isolation-by-distance pattern. Limnol Oceanogr 52:676–684
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
Ketmaier V, Mandatori R, De Matthaeis E, Mura G (2005) Molecular systematics and phylogeography in the fairy shrimp Tanymastix stagnalis based on mitochondrial DNA. J Zool 266:401–410
Ketmaier V, Pirollo D, De Matthaeis E, Tiedemann R, Mura G (2008) Large-scale mitochondrial phylogeography in the halophilic fairy shrimp Phallocryptus spinosa (Milne-Edwards 1840) (Branchiopoda: Anostraca). Aquat Sci 70:65–76
Kohn MH, York EC, Kamradt DA, Haught G, Sauvajot RM, Wayne RK (1999) Estimating population size by genotyping feces. Proc R Soc Lond B 266:657–663
Munoz J, Gomez A, Green AJ, Figuerola J, Amat F, Rico C (2008) Phylogeography and local endemism of the native Mediterranean brine shrimp Artemia salina (Branchiopoda:Anostraca). Mol Ecol 17:3160–3177
Nylander JAA (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Simovich MA, Hathaway SA (1997) Diversified bet-hedging as a reproductive strategy for some ephemeral vernal pool anostracans (Branchiopoda). J Crustac Biol 17:38–44
United States Fish and Wildlife Service (1994) Endangered and threatened wildlife and plants; determination of endangered status for the Conservancy fairy shrimp, longhorn fairy shrimp, and the vernal pool tadpole shrimp; and threatened status for the vernal pool fairy shrimp, vol 59, pp 48136–48153 (Federal Register)
Acknowledgments
I am grateful to R. Van Syoc (California Academy of Sciences) for access to the Branchinecta collections. I thank J. Baumsteiger, K. Kamer, and two anonymous reviewers for comments that improved the manuscript, as well as D.C. Rogers for discussion. D. Cooper provided valuable laboratory assistance. This work was supported by funds from the UC Merced Graduate Research Council. I also thank the US Fish and Wildlife Service (permit # TE-195305-0) California Department of Fish and Game (permit # SC-007388) for permission to collect B. lynchi.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Aguilar, A. Weak phylogeographic structure in the endemic western North American fairy shrimp Branchinecta lynchi (Eng, Belk and Erickson 1990). Aquat Sci 73, 15–20 (2011). https://doi.org/10.1007/s00027-010-0155-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00027-010-0155-6