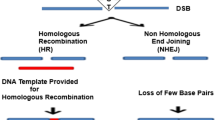
Abstract
Targeted genome modifications using techniques that alter the genomic information of interest have contributed to multiple studies in both basic and applied biology. Traditionally, in gene targeting, the target-site integration of a targeting vector by homologous recombination is used. However, this strategy has several technical problems. The first problem is the extremely low frequency of gene targeting, which makes obtaining recombinant clones an extremely labor intensive task. The second issue is the limited number of biomaterials to which gene targeting can be applied. Traditional gene targeting hardly occurs in most of the human adherent cell lines. However, a new approach using designer nucleases that can introduce site-specific double-strand breaks in genomic DNAs has increased the efficiency of gene targeting. This new method has also expanded the number of biomaterials to which gene targeting could be applied. Here, we summarize various strategies for target gene modification, including a comparison of traditional gene targeting with designer nucleases.





Similar content being viewed by others
References
Madyagol M, Al-Alami H, Levarski Z, Drahovska H, Turna J, Stuchlik S (2011) Gene replacement techniques for Escherichia coli genome modification. Folia Microbiol Praha 56(3):253–263. doi:10.1007/s12223-011-0035-z
Bouabe H, Okkenhaug K (2013) Gene targeting in mice: a review. Methods Mol Biol 1064:315–336. doi:10.1007/978-1-62703-601-6_23
Puchta H, Fauser F (2013) Gene targeting in plants: 25 years later. Int J Dev Biol 57(6–8):629–637. doi:10.1387/ijdb.130194hp
Maggio I, Goncalves MA (2015) Genome editing at the crossroads of delivery, specificity, and fidelity. Trends Biotechnol 33(5):280–291. doi:10.1016/j.tibtech.2015.02.011
Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S (2001) Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol 21(8):2858–2866. doi:10.1128/MCB.21.8.2858-2866.2001
Langston LD, Symington LS (2004) Gene targeting in yeast is initiated by two independent strand invasions. Proc Natl Acad Sci USA 101(43):15392–15397. doi:10.1073/pnas.0403748101
Yamazoe M, Sonoda E, Hochegger H, Takeda S (2004) Reverse genetic studies of the DNA damage response in the chicken B lymphocyte line DT40. DNA Repair Amst 3(8–9):1175–1185. doi:10.1016/j.dnarep.2004.03.039
Abuin A, Zhang H, Bradley A (2000) Genetic analysis of mouse embryonic stem cells bearing Msh3 and Msh2 single and compound mutations. Mol Cell Biol 20(1):149–157
Kuhn R, Schwenk F (1997) Advances in gene targeting methods. Curr Opin Immunol 9(2):183–188
Agarwal S, van Cappellen WA, Guenole A, Eppink B, Linsen SE, Meijering E, Houtsmuller A, Kanaar R, Essers J (2011) ATP-dependent and independent functions of Rad54 in genome maintenance. J Cell Biol 192(5):735–750. doi:10.1083/jcb.201011025
Munoz IG, Prieto J, Subramanian S, Coloma J, Redondo P, Villate M, Merino N, Marenchino M, D’Abramo M, Gervasio FL, Grizot S, Daboussi F, Smith J, Chion-Sotinel I, Paques F, Duchateau P, Alibes A, Stricher F, Serrano L, Blanco FJ, Montoya G (2011) Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus. Nucleic Acids Res 39(2):729–743. doi:10.1093/nar/gkq801
Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39(12):e82. doi:10.1093/nar/gkr218
Yanez RJ, Porter AC (1998) Therapeutic gene targeting. Gene Ther 5(2):149–159. doi:10.1038/sj.gt.3300601
Adachi N, Nishijima H, Shibahara K (2008) Gene targeting using the human Nalm-6 pre-B cell line. Biosci Trends 2(5):169–180
Jasin M, Rothstein R (2013) Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol 5(11):a012740. doi:10.1101/cshperspect.a012740
Wesoly J, Agarwal S, Sigurdsson S, Bussen W, Van Komen S, Qin J, van Steeg H, van Benthem J, Wassenaar E, Baarends WM, Ghazvini M, Tafel AA, Heath H, Galjart N, Essers J, Grootegoed JA, Arnheim N, Bezzubova O, Buerstedde JM, Sung P, Kanaar R (2006) Differential contributions of mammalian Rad54 paralogs to recombination, DNA damage repair, and meiosis. Mol Cell Biol 26(3):976–989. doi:10.1128/MCB.26.3.976-989.2006
Ferretti LP, Lafranchi L, Sartori AA (2013) Controlling DNA-end resection: a new task for CDKs. Front Genet 4:99. doi:10.3389/fgene.2013.00099
Motycka TA, Bessho T, Post SM, Sung P, Tomkinson AE (2004) Physical and functional interaction between the XPF/ERCC1 endonuclease and hRad52. J Biol Chem 279(14):13634–13639. doi:10.1074/jbc.M313779200
Huhn D, Bolck HA, Sartori AA (2013) Targeting DNA double-strand break signalling and repair: recent advances in cancer therapy. Swiss Med Wkly 143:w13837. doi:10.4414/smw.2013.13837
Baumann P, Benson FE, West SC (1996) Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell 87(4):757–766
van Veelen LR, Essers J, van de Rakt MW, Odijk H, Pastink A, Zdzienicka MZ, Paulusma CC, Kanaar R (2005) Ionizing radiation-induced foci formation of mammalian Rad51 and Rad54 depends on the Rad51 paralogs, but not on Rad52. Mutat Res 574(1–2):34–49. doi:10.1016/j.mrfmmm.2005.01.020
Andersen SL, Sekelsky J (2010) Meiotic versus mitotic recombination: two different routes for double-strand break repair: the different functions of meiotic versus mitotic DSB repair are reflected in different pathway usage and different outcomes. BioEssays 32(12):1058–1066. doi:10.1002/bies.201000087
Luo G, Santoro IM, McDaniel LD, Nishijima I, Mills M, Youssoufian H, Vogel H, Schultz RA, Bradley A (2000) Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat Genet 26(4):424–429. doi:10.1038/82548
Yamanishi A, Yusa K, Horie K, Tokunaga M, Kusano K, Kokubu C, Takeda J (2013) Enhancement of microhomology-mediated genomic rearrangements by transient loss of mouse Bloom syndrome helicase. Genome Res 23(9):1462–1473. doi:10.1101/gr.152744.112
Sarbajna S, West SC (2014) Holliday junction processing enzymes as guardians of genome stability. Trends Biochem Sci 39(9):409–419. doi:10.1016/j.tibs.2014.07.003
Chen XB, Melchionna R, Denis CM, Gaillard PH, Blasina A, Van de Weyer I, Boddy MN, Russell P, Vialard J, McGowan CH (2001) Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol Cell 8(5):1117–1127
Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR 3rd, Russell P, Fuchs RP, McGowan CH, Gaillard PH (2009) Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138(1):78–89. doi:10.1016/j.cell.2009.06.029
Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, Elledge SJ, Harper JW (2009) Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138(1):63–77. doi:10.1016/j.cell.2009.06.030
Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC (2008) Identification of Holliday junction resolvases from humans and yeast. Nature 456(7220):357–361. doi:10.1038/nature07470
Niedernhofer LJ, Essers J, Weeda G, Beverloo B, de Wit J, Muijtjens M, Odijk H, Hoeijmakers JH, Kanaar R (2001) The structure-specific endonuclease Ercc1–Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J 20(22):6540–6549. doi:10.1093/emboj/20.22.6540
Abraham J, Lemmers B, Hande MP, Moynahan ME, Chahwan C, Ciccia A, Essers J, Hanada K, Chahwan R, Khaw AK, McPherson P, Shehabeldin A, Laister R, Arrowsmith C, Kanaar R, West SC, Jasin M, Hakem R (2003) Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells. EMBO J 22(22):6137–6147. doi:10.1093/emboj/cdg580
McPherson JP, Lemmers B, Chahwan R, Pamidi A, Migon E, Matysiak-Zablocki E, Moynahan ME, Essers J, Hanada K, Poonepalli A, Sanchez-Sweatman O, Khokha R, Kanaar R, Jasin M, Hande MP, Hakem R (2004) Involvement of mammalian Mus81 in genome integrity and tumor suppression. Science 304(5678):1822–1826. doi:10.1126/science.1094557
Rijkers T, Van Den Ouweland J, Morolli B, Rolink AG, Baarends WM, Van Sloun PP, Lohman PH, Pastink A (1998) Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol Cell Biol 18(11):6423–6429
Nakahara M, Sonoda E, Nojima K, Sale JE, Takenaka K, Kikuchi K, Taniguchi Y, Nakamura K, Sumitomo Y, Bree RT, Lowndes NF, Takeda S (2009) Genetic evidence for single-strand lesions initiating Nbs1-dependent homologous recombination in diversification of Ig v in chicken B lymphocytes. PLoS Genet 5(1):e1000356. doi:10.1371/journal.pgen.1000356
Langston LD, Symington LS (2005) Opposing roles for DNA structure-specific proteins Rad1, Msh2, Msh3, and Sgs1 in yeast gene targeting. EMBO J 24(12):2214–2223. doi:10.1038/sj.emboj.7600698
Menke DB (2013) Engineering subtle targeted mutations into the mouse genome. Genesis 51(9):605–618. doi:10.1002/dvg.22422
Lin SC, Chang YY, Chan CC (2014) Strategies for gene disruption in Drosophila. Cell Biosci 4(1):63. doi:10.1186/2045-3701-4-63
Gonzales AP, Yeh JR (2014) Cas9-based genome editing in zebrafish. Methods Enzymol 546:377–413. doi:10.1016/B978-0-12-801185-0.00018-0
Van Nieuwenhuysen T, Naert T, Tran HT, Van Imschoot G, Geurs S, Sanders E, Creytens D, Van Roy F, Vleminckx K (2015) TALEN-mediated apc mutation in Xenopus tropicalis phenocopies familial adenomatous polyposis. Oncoscience 2(5):555–566
Nakayama T, Blitz IL, Fish MB, Odeleye AO, Manohar S, Cho KW, Grainger RM (2014) Cas9-based genome editing in Xenopus tropicalis. Methods Enzymol 546:355–375. doi:10.1016/B978-0-12-801185-0.00017-9
Mashimo T (2014) Gene targeting technologies in rats: zinc finger nucleases, transcription activator-like effector nucleases, and clustered regularly interspaced short palindromic repeats. Dev Growth Differ 56(1):46–52. doi:10.1111/dgd.12110
Honda A, Hirose M, Sankai T, Yasmin L, Yuzawa K, Honsho K, Izu H, Iguchi A, Ikawa M, Ogura A (2015) Single-step generation of rabbits carrying a targeted allele of the tyrosinase gene using CRISPR/cas9. Exp Anim 64(1):31–37. doi:10.1538/expanim.14-0034
Wu H, Wang Y, Zhang Y, Yang M, Lv J, Liu J, Zhang Y (2015) TALE nickase-mediated SP110 knockin endows cattle with increased resistance to tuberculosis. Proc Natl Acad Sci USA 112(13):E1530–E1539. doi:10.1073/pnas.1421587112
Pabo CO, Peisach E, Grant RA (2001) Design and selection of novel Cys2His2 zinc finger proteins. Annu Rev Biochem 70:313–340. doi:10.1146/annurev.biochem.70.1.313
Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S (2005) Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res 33(18):5978–5990. doi:10.1093/nar/gki912
Gaj T, Gersbach CA, Barbas CF 3rd (2013) ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31(7):397–405. doi:10.1016/j.tibtech.2013.04.004
Fu F, Voytas DF (2013) Zinc Finger Database (ZiFDB) v2.0: a comprehensive database of C(2)H(2) zinc fingers and engineered zinc finger arrays. Nucleic Acids Res 41(Database issue):D452–D455. doi:10.1093/nar/gks1167
Gupta A, Meng X, Zhu LJ, Lawson ND, Wolfe SA (2011) Zinc finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc finger nucleases. Nucleic Acids Res 39(1):381–392. doi:10.1093/nar/gkq787
Boch J, Bonas U (2010) Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol 48:419–436. doi:10.1146/annurev-phyto-080508-081936
Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF (2010) Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186(2):757–761. doi:10.1534/genetics.110.120717
Streubel J, Blucher C, Landgraf A, Boch J (2012) TAL effector RVD specificities and efficiencies. Nat Biotechnol 30(7):593–595. doi:10.1038/nbt.2304
Li K, Wang G, Andersen T, Zhou P, Pu WT (2014) Optimization of genome engineering approaches with the CRISPR/cas9 system. PLoS One 9(8):e105779. doi:10.1371/journal.pone.0105779
Seligman LM, Chisholm KM, Chevalier BS, Chadsey MS, Edwards ST, Savage JH, Veillet AL (2002) Mutations altering the cleavage specificity of a homing endonuclease. Nucleic Acids Res 30(17):3870–3879
Epinat JC, Arnould S, Chames P, Rochaix P, Desfontaines D, Puzin C, Patin A, Zanghellini A, Paques F, Lacroix E (2003) A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res 31(11):2952–2962
Rosen LE, Morrison HA, Masri S, Brown MJ, Springstubb B, Sussman D, Stoddard BL, Seligman LM (2006) Homing endonuclease I-CreI derivatives with novel DNA target specificities. Nucleic Acids Res 34(17):4791–4800. doi:10.1093/nar/gkl645
Arnould S, Perez C, Cabaniols JP, Smith J, Gouble A, Grizot S, Epinat JC, Duclert A, Duchateau P, Paques F (2007) Engineered I-CreI derivatives cleaving sequences from the human XPC gene can induce highly efficient gene correction in mammalian cells. J Mol Biol 371(1):49–65. doi:10.1016/j.jmb.2007.04.079
Grizot S, Smith J, Daboussi F, Prieto J, Redondo P, Merino N, Villate M, Thomas S, Lemaire L, Montoya G, Blanco FJ, Paques F, Duchateau P (2009) Efficient targeting of a SCID gene by an engineered single-chain homing endonuclease. Nucleic Acids Res 37(16):5405–5419. doi:10.1093/nar/gkp548
Saleh-Gohari N, Helleday T (2004) Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res 32(12):3683–3688. doi:10.1093/nar/gkh703
Ozeri-Galai E, Tur-Sinai M, Bester AC, Kerem B (2014) Interplay between genetic and epigenetic factors governs common fragile site instability in cancer. Cell Mol Life Sci 71(23):4495–4506. doi:10.1007/s00018-014-1719-8
Gottipati P, Helleday T (2009) Transcription-associated recombination in eukaryotes: link between transcription, replication and recombination. Mutagenesis 24(3):203–210. doi:10.1093/mutage/gen072
Acknowledgments
KH is funded by a Grant-in-Aid for Young Scientists (A) (25710010), Japan Society for the Promotion of Science (JSPS), The Ministry of Education, Culture, Sports, Science and Technology (MEXT). The authors declare that no conflict of interest exists.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tokunaga, A., Anai, H. & Hanada, K. Mechanisms of gene targeting in higher eukaryotes. Cell. Mol. Life Sci. 73, 523–533 (2016). https://doi.org/10.1007/s00018-015-2073-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-015-2073-1