Abstract
MicroRNAs (miRNAs) exhibit many functions in biological activities. Recent studies have shown that miRNAs exist outside cells and are transferred between cells. Extracellular miRNAs are protected from ribonucleases found in body fluids through binding to specific proteins or by being encapsulated in lipid bilayer vesicles. Here, we review the mechanisms of the secretion and uptake as well as the functions of extracellular miRNAs, particularly those encapsulated in extracellular vesicles. Extracellular vesicles are related to cancer progression, and some miRNAs in extracellular vesicles are associated with cancer cells. We describe the transfer of cancer-related miRNAs between cancer cells and non-cancerous cells. Finally, we discuss the anticipated applications of miRNAs present in extracellular vesicles in diagnostics and therapeutics.
Similar content being viewed by others
Introduction
MicroRNAs (miRNAs) are small (18–25 nt) non-coding RNAs that repress gene expression through post-transcriptional degradation or translational repression [1]. The first identified miRNA, lin-4, was found in Caenorhabditis elegans in 1993 and was suggested to regulate transcription via an RNA–RNA interaction in the 3′ untranslated region (UTR) of lin-14 mRNA [2]. miRNAs are conserved in a large number of species of living organisms [3, 4]. So far, 2,588 miRNAs have been identified in Homo sapiens [5].
The most fundamental function of miRNAs is RNA interference (RNAi), which is a mechanism of silencing gene expression by degrading mRNAs with a complementary sequence. The phenomenon of silencing by small RNA fragments was first observed in petunia plants in 1990 (referred to as ‘cosuppression’) [6, 7] and subsequently in Neurospora crassa in 1992 (referred to as ‘quelling’) [8], but the underlying molecular mechanism was unclear. In 1998, Mello and Fire first described the RNAi mechanism in studies on C. elegans using short interfering RNAs (siRNAs) [9, 10]. Since then, RNAi has been shown to be conserved in almost all species and to possess a common, fundamental mechanism of regulating gene expression. The discovery of RNAi has also accelerated molecular biology research by leading to the development of genetic technologies to knock down any desired gene.
The molecular mechanism of RNAi has been explained in detail previously (reviewed in [11]). In brief, a miRNA is processed from a stem-loop sequence of ~70 nt (pre-mir), which is excised from a longer primary transcript (pri-mir). The pre-mir is exported to the cytoplasm to be cleaved into double-stranded RNA to form the miRNA by Dicer, an RNase III enzyme. The double-stranded miRNA is incorporated into the miRISC complex, and the leading strand of the miRNA is retained in the complex. miRNAs bind to a perfect complementary or a nearly perfect complementary sequence within a mRNA, and the sequence is cleaved by the miRISC complex.
Our knowledge of the importance of miRNAs in various bioactivities has been increasing. In addition to RNAi, miRNA regulates transcriptional levels in another manner: through inhibition of translation by binding to the 3′-UTR [12] and 5′-UTR of mRNAs [13]. Recent studies have also shown that miRNAs recognize coding regions [14]. Because miRNAs recognize sequences with imperfect pairings, a single miRNA can target a broad range of mRNAs with nearly complementary sequences, thereby increasing its impact on transcriptional regulation. Furthermore, miRNAs are involved in gene silencing through chromatin remodeling [15], which implies that miRNAs also play a role in epigenetic regulation. miRNAs have also been suggested to regulate DNA methylation and telomere recombination [16]. Through these mechanisms, miRNAs are involved in many physiological roles and developmental processes as well as in the progression of some diseases, such as cancer [17]. A reduction of tumor-suppressive miRNAs or overexpression of tumor-promoting miRNAs has been observed in cancer progression [18].
These post-transcriptional regulation and the other functions of miRNAs are all carried out in the cytoplasm or nuclear envelope; that is, inside the cell. However, miRNAs have also been found outside cells in various body fluids, such as serum, saliva, urine, breast milk, and tears [19–22]. These miRNAs are spontaneously secreted from cells. Moreover, these extracellular miRNAs function in recipient cells, indicating roles in cell–cell communication. Extracellular miRNAs also have been suggested to be related to several diseases, including cancer. In this review, we describe cell–cell communication between cancer cells and non-cancerous cells via miRNA packaged in extracellular vesicles and potential applications for therapeutic strategies.
Extracellular miRNAs
RNAs have been discovered outside cells, even though RNAs have generally been thought to function inside cells. The following types of RNA have been found in extracellular environments: mRNA, tRNA, miRNA, small interfering RNA (siRNA), and lncRNA; however, almost no rRNA was detected in studies on a mast cell line and glioblastoma [23, 24]. In humans, miRNAs have been found in several body fluids, such as blood plasma [22], urine [25], saliva [26], semen [27] breast milk [28], and cerebrospinal fluid [29]. The detection of miRNAs outside cells drew interest because some body fluids, such as saliva, milk, and urine, contain a high concentration of nucleases [30]. Indeed, miRNAs from serum were shown to remain stable after several treatments that degraded most RNAs [31]. These reports suggested that extracellular miRNAs are protected from ribonucleases in some manner. Several mechanisms for preventing the degradation of miRNAs have been demonstrated. One possible mechanism used to protect miRNAs from degradation is RNA-binding proteins, such as Argonaute 2 (Ago2) [32], other Ago proteins (Ago1 [33], Ago3, Ago4 [34]), and nucleophosmin 1 [35]. High-density lipoproteins (HDLs) and low-density lipoproteins (LDLs) also interact with and transfer miRNAs to cells [36]. An alternative mechanism used to protect miRNAs from degradation involves the packaging of miRNAs in small, lipid bilayer vesicles, referred to as extracellular vesicles (EVs) [23]. Previously, we have found that approximately 30 % of extracellular miRNAs are packaged into EVs in both cell culture and sera (unpublished). These two mechanisms are independent according to a study showing that Ago2 transfers miRNAs independent of EVs based on differential centrifugation and size-exclusion chromatography analyses [32]. However, some reports discuss the possibility that extracellular miRNAs associated with proteins are not spontaneously secreted but represent cellular residue released from dead cells [34]. In addition, evidence of the uptake of protein-associated miRNAs and their function in the recipient cells remains scarce. The mechanisms and functions of protein-bound miRNAs should be elucidated to demonstrate the biological role of miRNAs associated with protein and to determine whether extracellular miRNAs protected by proteins and miRNAs encapsulated in EVs exhibit different secretion pathways and functions. This review primarily addresses miRNAs secreted in vesicular form, for which the mechanism has been more clearly elucidated.
Extracellular vesicles (EVs)
Extracellular vesicle is a comprehensive term for lipid bilayered vesicles that exist outside cells [37]. EVs include apoptotic bodies (800–5,000 nm diameter), microvesicles (50–1,000 nm diameter), and exosomes (40–100 nm diameter) [38, 39]. These EVs have different origins and properties. Apoptotic bodies are released from the cell surface during the process of apoptosis [40]. Microvesicles are also formed by budding directly from the cellular membrane [41], whereas exosomes first bud into a multivesicular body (MVB), which is an intracellular compartment, and are then secreted to the extracellular milieu [39]. Different RNA profiles have been demonstrated for each type of vesicle [42]. Apoptotic bodies primarily contain rRNA, and microvesicles carry little RNA, except for microvesicles from TF-1 cell culture. Exosomes contain mRNA and miRNA, but little rRNA [42]. Proteins on the surface are also different among the three vesicles. CD9 and CD63 are recognized as potential markers of exosomes, ARF6 and VCAMP2 are proposed as microvesicle markers, and TSP and C3b are accepted as markers for apoptotic bodies (reviewed in [43]). With respect to lipid compositions, exosomes have low phosphatidylserine (PS) at outer leaflet, while microvesicles and apoptotic bodies contain high amount of PS [44]. Enrichment of ceramide is one of the characteristics of exosomes [45].
However, we must consider that the currently used techniques are inadequate for collecting each type of EV separately [46]. In addition, the nomenclature of exosomes is confusing [37]. In 1983, two groups reported that reticulocytes release transferrin receptors associated with small vesicles [47, 48]. These authors showed that the vesicles were formed inside multivesicular endosomes. The term ‘exosome’ was subsequently applied to the vesicles in 1987 [49]. However, in 1981, microvesicles released to cell cultures from the cell membrane of normal and neoplastic cell lines with 5′-nucleotidase activity were referred to as ‘exosomes’ [50]. ‘Exosome’ is also used to refer to the exoribonucleases identified in Saccharomyces cerevisiae [51]. The International Society of Extracellular Vesicles recommended the nomenclature ‘EVs’ as a generic term for all secreted vesicles, and the terms should be defined clearly before use [37]. In this review, we use the term ‘EVs’ for collective extracellular vesicles, and ‘exosomes’ for extracellular vesicles which are formed by budding into MVBs.
EVs have long been thought of as no more than a garbage can for cells to discard unwanted components [52]. mRNAs, DNAs, and proteins have been detected within these vesicles [53], and miRNAs were discovered inside EVs decades later, in 2007 [23]. Valadi et al. identified miRNAs as well as mRNAs inside EVs from mouse and human mast cell lines and proved that these nucleic acids are functional in the recipient cells. These findings drew much attention to EVs, and miRNAs were recognized as a new regulatory component in gene expression. Based on the reports accumulated to date, almost all cells are now considered to release EVs [23].
Secretion of miRNAs packed in exosomes
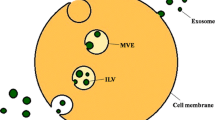
Studies examining the molecular mechanisms of exosomes release from one cell and their uptake by other cells are ongoing (described in Fig. 1). Exosomes are formed by budding into early endosomes to form MVBs. MVBs fuse with the cellular membrane or lysosomes. When fused with the plasma membrane, internal exosomes are released outside the cell [54].
Schematic representation of the secretion, transfer, and uptake of exosomes. Exosomes are formed by budding into early endosomes to form MVBs. The internal vesicles are secreted outside the cells via MVB fusion with the cellular membrane. The released exosomes are taken up by neighboring cells or distant cells. Exosome internalization occurs through either direct fusion or endocytic pathways. The exosomes are carried outside the cells for lipid recycling
The mechanism underlying the formation of exosomes into MVBs is controversial. The biogenesis of exosomes was first suggested to be associated with the endosomal sorting complex required for transport (ESCRT) [55]. ESCRT-related proteins, such as HRS (ESCRT-0), TSG101 (ESCRT-I), CHMP4 (ESCRT-II), ALIX and VPS4 (ESCRT-III), have been shown to be recruited to the cytosolic sides of MVBs to form exosomes (summarized in [56]). However, a recent report demonstrated that mammalian cells without key ESCRT components could still form MVBs [57]. Some studies have revealed some exosome formation that is independent of ESCRT, and this process depends on several molecules, such as ceramide [45] or tetraspanins [58]. Neutral sphingomyelinase 2 (nSMase2), a rate-limiting enzyme in ceramide biosynthesis [59], affects the secretion of miRNAs [60]. The nSMase2-dependent transfer of miRNAs via exosomes contributes to cancer metastasis in mouse breast cancer [59]. Tetraspanins are enriched on the surface of exosomes, and some of these proteins, such as CD9, CD63, and CD81, are also used as marker proteins of the exosome. Bone marrow dendritic cells from CD9 knockout mice were incapable to secrete exosome [61], while CD81 seemed not effect exosome secretion in lymphocytes [62] (reviewed in [58]). CD63 has been suggested to be instrumental in the formation of internal vesicles in melanocytes [63].
MVBs are sorted in two ways: through fusion with lysosomes or through fusion with the cellular membrane to secrete exosomes [56]. During the secretion steps, RAB proteins (RAB11, RAB27, and RAB35) have been shown to be involved in the transport of MVBs to the plasma membrane and in exosome secretion [56]. In addition, SNAREs have been suggested to be involved in MVB fusion with the plasma membrane [46].
Within exosomes from human, more than 700 miRNAs have been identified to date, together with thousands of proteins, DNAs, and mRNAs (Vesiclepedia and EVpedia) [64–67]. Several studies using microarrays or next-generation sequencing have identified a dozens of non-coding RNAs in exosomes froma single cell line [68, 69]. However, based on the size of exosomes (~100 nm), not all miRNAs are packaged into a single exosome. Exosomes from the same cell line are likely to exhibit different contents [70, 71].
The miRNA concentration within exosomes appears to partially depend on the concentration inside the cell. For example, exosomes secreted from cells overexpressing a certain miRNAs contain more of the overexpressed miRNAs [72]. However, many reports have shown that the profiles of miRNAs in exosomes are different from those of exosome-producing cells, suggesting that miRNAs are selectively packaged into exosomes. For example, although rRNA accounts for the majority of the RNA in a cell, almost no rRNA was detected in studies examining exosomes from a mast cell line and glioblastoma [23, 24]. The first paper detecting miRNAs in exosomes showed that only a fraction of miRNAs from all miRNAs expressed in the cells were detected in exosomes [23]. Moreover, some miRNAs were found at higher levels in exosomes than in the cells [23]. In MCF7 cells (breast cancer cells), only 2 % of many miRNAs including miR-720, the most abundant miRNA in the cell, were released into the environment, whereas miR-451 and miR-107 were secreted at higher levels [73]. However, the molecules responsible for selectively packaging miRNAs into exosomes remain to be identified. Villarroya-Beltri et al. [74] discussed that sumoylated form of the heterogenous ribonucleoprotein A2B1 (hnRNPA2B1) would load miRNAs into exosomes through EXOmotif (GGAG) in the miRNAs. The mechanisms of miRNA secretion can be applied to design exosomes that contain specific miRNAs. This technique would enable us to produce exosomes carrying tumor-suppressive miRNAs for cancer therapy.
Uptake of exosomes into target cells
Released exosomes may be taken up by nearby cells, or they may reach cells at a distance through the blood or lymph system. However, the mechanism of uptake is more obscure than the secretion mechanism. The mechanisms of uptake suggested to date include endocytic pathways, such as endocytosis, phagocytosis and macropinocytosis, and direct fusion with the cellular membrane [54, 75–78] (Fig. 1). Escrevente et al. [79] showed that proteinase K treatment of exosomes significantly reduced uptake by ovarian cancer cells. The results indicate that surface proteins on exosomes take part in the uptake of exosomes. Mulcahy et al. suggested in a review that proteins on the surface of exosomes such as tetraspanins and lectins are involved in exosome uptake. They also suggested that integrins, proteoglycans, and lectin receptor proteins on the cell surface may serve as receptor of exosome surface protein for uptake (reviewed in [80]).
Concerning the direct fusion mechanism, Parolini et al. showed, using fluorescently labeled exosomes from melanoma cells, that lower pH, which is linked to tumor malignancy, increases exosome release and uptake. The authors indicated that a high rigidity and a high content of sphingomyelin/ganglioside GM3 and caveolin-1 in exosomes are responsible for accelerated fusion under a low pH [78].
Endocytic uptake was first suggested in dendritic cells [81]. Some reports have identified the molecules involved in exosome uptake through endocytosis. Dynamin 2 (Dyn2), which is necessary for clathrin- and caveolin-dependent endocytosis, was shown to be essential for the internalization of exosomes, suggesting that exosomes are taken up via clathrin- or caveolin-dependent endocytosis [75]. Heparan sulfate proteoglycans (HSPGs) have been suggested to serve as the receptors of cancer cell-derived exosomes in glioblastoma patients [82]. This finding suggests a mechanism similar to a virus entering a cell, as HSPGs are also known to be involved in viral entry [82]. In addition, exosomes from glioblastoma cells are internalized into HUVECs through lipid raft-mediated endocytosis and are negatively regulated by caveolin-1 (CAV1), although CAV1 was not found to co-localize with exosomes. CAV1 was suggested to have negative effect on exosome internalization through extracellular signal-regulated kinase-1/2 (ERK1/2)- and heat shock protein 27 (HSP27) signaling [83].
Tian et al. [84] visualized the cellular uptake and intracellular trafficking of exosomes in vitro at a single-particle level and indicated that exosomes may enter cells through endocytic pathways. First, exosomes are transported along the cell periphery, potentially by actin filaments. Exosomes are then carried to perinuclear regions, possibly on microtubules via dynein-dependent transport, to be trapped in large vesicles. At the same time, exosomes are transferred from the perinuclear space to the cell surface, which may occur along microtubules via kinesins for lipid recycling. The proteins in exosomes were observed to be released from exosomes within 3 h after endocytosis and through the fusion of endosomes or late endosomes in an acidified environment. Then, the proteins are sorted into lysosomes. When and how miRNAs are released to functionally regulate transcription in the cytosol has not been demonstrated.
Whether exosomes are selectively taken up by certain cells has been investigated. Many reports have suggested that exosomes are delivered specifically to cells [85]. Unidirectional transfer of miRNAs from T cells to antigen-presenting cells via exosomes has been reported [72]. Proteins at the surface of exosomes seemed to be involved in target-specific internalization into cells. However, the primary mechanism by which exosomes selectively interact with target cells has not been elucidated.
After exosomes are uptaken by recipient cells, exosome-transferred miRNAs function in transcriptional regulation in recipient cells [53]. In 2010, four independent groups showed that miRNAs transferred to recipient cells function to repress the expression of target genes [60, 86–88]. As exosomes are thought to be secreted from all types of cells, these vesicles are likely to be responsible for important physiological functions through cell–cell communication. Indeed, reports have shown that exosomes are involved in physiological functions, for example, in immune response (T cells, B cell, and dendritic immune cells) [28, 72] and neuron-glia communication [89]. However, the molecular mechanisms of how exosomes act in physiological functions have remained elusive because of the limits of the experimental methods for detection and for tracking exosomes from specific cells in vivo.
Cancer-promoting EVs and their miRNAs
The expression levels of miRNAs are dynamically altered in cancer cells compared to normal cells [86]. Many miRNAs have been reported to play roles in both accelerating and inhibiting of cancer progression [20, 87]. These cancer-related miRNAs have been detected not only in cancer cells but also in cancer-derived EVs (reviewed in [52]). Indeed, increasing evidences suggested that EVs contribute to malignancy in cancer [90]. Cancer cells release more EVs than normal cells [91, 92]. Tumor-derived EVs are recognized as antigens by dendritic cells, inducing antitumor immune responses [70]; at the same time, cancer-derived EVs are associated with tumor malignancy and progression. Inhibition of EV secretion using a short hairpin RNA targeting RAB27A, a protein involved in intracellular trafficking of EVs, led to poor development of subcutaneously injected 4T1 mouse mammary tumor cells [93]. Furthermore, inhibition of EV secretion through nSMase2 knockdown inhibited vascular formation in primary tumors, and the opposite response was observed after nSMase2 overexpression [59]. Here, we describe some of the cancer progression-related miRNAs detected in EVs thus far (Table 1).
Tumor proliferation and angiogenesis
Communication between tumor cells and their surrounding microenvironment has a large impact on tumor proliferation [94]. Cancer cells modify their microenvironment by transferring miRNAs that are suitable for their growth via EVs. miR-210 is a major cancer-related miRNA that is induced in hypoxia [95]. In EVs from highly metastatic breast cancer, miR-210 was found to be enriched and to be responsible for the induction of angiogenesis in the tumor microenvironment [59]. Endothelial cells that received tumor-derived EVs underwent vascular formation [59]. miR-9 is delivered by melanoma-derived EVs into endothelial cells and promotes endothelial cell migration and angiogenesis by reducing SOCS5 and activating the JAK-STAT pathway [96]. miR-105 in EVs from breast cancer cells targets the tight junction protein ZO-1, leading to destruction of vascular endothelial barriers and promotes metastasis [97].
Cancer cells release miRNAs to decrease the expression of tumor suppressor genes. miR-21, which contributes to the development of cancer, has also been identified in cancer-derived EVs [98]. miR-21 represses PTEN [99] and PDCD4 [100], which are known tumor suppressor genes. miR-21 has been detected in EVs from breast cancer cells [73], glioblastoma [98], and malignant ascites [101]. Let-7 miRNA, which was found to play tumor-suppressive roles in early studies [102], accumulates in secreted EVs from a metastatic gastric cancer cell line, AZ-P7a, compared with low-metastatic cells and other types of cancer cell lines [103]. Let-7 miRNA downregulates oncogenes such as RAS and HMGA2, suggesting that AZ-P7a cells release let-7 miRNA via EVs to maintain their oncogenesis [103].
Tumor dormancy
Breast cancer is known to present especially late recurrence after 10–20 years [104]. EVs from bone marrow mesenchymal stem cells (BM-MSCs) are thought to enable breast cancer cells in the bone marrow to be dormant for 10 years, which is considered the main cause of breast cancer reoccurrence after a long period [105]. miR-23b in EVs from BM-MSCs were suggested to suppress MARCS, which encodes myristoylated alanine-rich C kinase substrate and promotes cell cycling and motility [105].
Transfer of drug resistance
In cancer, EVs also contribute to malignancy by enhancing the drug resistance of the recipient cells. Using the MCF-7 cell line, Chen et al. showed that EVs from drug-resistant breast cancer cells enhance drug resistance in proximate cells by modulating the cell cycle and drug-induced apoptosis. Through microarray analyses and confirmation via quantitative real-time PCR, miR-100, miR-222, and miR-30a were shown to be increased in EVs from drug-resistant breast cancer cells, suggesting that these miRNAs contribute to the transfer of drug resistance [106].
Suppression of the immune system
During cancer progression, the immune system attempts to remove cancer cells. However, many reports have shown that EVs released from cancer cells can repress the immune system through multiple pathways [107, 108]. In most cases, proteins within and on the surface of EVs play roles in suppression; however, miR-21 and miR-29a in EVs derived from lung cancer cells were observed to be able to bind to Toll-like receptors on surrounding immune cells to trigger an inflammatory response, leading to the secretion of pro-metastatic inflammatory cytokines [109]. Cancer cells also take advantage of immune cell-derived EVs. Tumor-associated macrophages were shown to release EVs containing miR-223, which is specific for IL-4-activated M2 macrophages. miR-223 transferred to breast cancer cells targets the 3′-UTR of MEF2C, a myocyte enhancer factor. The reduction of MEF2C is linked to nuclear accumulation of the function of β-catenin to promote the invasiveness of the SKBR3 and MDA-MB-231 breast cancer cell lines [110].
Tumor metastasis
Extracellular vesicles derived from cancer cells have also been suggested to contribute to metastasis by modifying the environment of distant organs. To form colonies in metastatic organs, cancer cells must overcome several barriers, one of which is surviving and proliferating in metastatic organs. Tumor cells have been suggested to condition the metastatic area beforehand using EVs to form an environment that is suitable for growing cancer cells. The microarea conditioned by EVs is referred to as the premetastatic niche [111]. Some miRNAs involved in the formation of the premetastatic niche have been identified. In renal cancer stem cells, several miRNAs (miR-200c, miR-92, miR-141, miR-29a, miR-650, and miR-151) found in EVs are related to the formation of the lung premetastatic niche and lead to tumor invasion and metastasis, in addition to inducing angiogenesis [112]. In EVs from metastatic rat adenocarcinoma, miR-494 and miR-542-3p were shown to be increased and to prepare premetastatic organ stroma cells by affecting proteases, adhesion molecules, chemokine ligands, cell cycle- and angiogenesis-promoting genes, and the oxidative stress response [113].
EVs from non-cancerous cells with tumor-suppressive effects
Many reports have shown that EVs from cancer cells contribute to cancer progression. However, normal cells also release EVs that play inhibitory roles in cancer progression, although few transferring tumor-suppressive miRNAs have been reported to inhibit the proliferation of cancer cells. Epithelial prostate cells, PNT-2, secrete EVs containing miR-143, which repressed proliferation of prostate cancer cells, PC-3M [114]. Other tumor-suppressive miRNAs, including miR-15a, the miR-200 family, miR-148a, miR-193b, miR-126, and miR-205, have also been identified. miR-143 downregulates KRAS and ERK5 and represses the proliferation of metastatic prostate cancer PC-3M cells [114]. Some reports have indicated that immune cells release tumor-suppressive EVs. For example, NK cells release EVs containing perforin, CD56, and granzyme B to inhibit tumor growth [115]. However, in these EVs, only proteins, and not miRNAs, have been identified as effective components thus far. However, it is likely that immune cell-derived EVs also contain miRNAs, which contribute to suppress tumor progression.
miRNAs in EVs as biomarkers for diagnostic, prognostic, and therapeutic targets
Extracellular miRNAs are anticipated to be useful as biomarkers because miRNAs in vesicles can be detected in body fluids (reviewed elsewhere, in [91, 116]), such as sera, urine, and saliva. In addition, EVs reflect different expression profiles in normal cells and cancerous cells in ovarian cancer [91], or cancer-derived exosomes selectively contain specific miRNAs in cells [73]. The profiling of miRNAs in EVs has become easier due to the development of microarray and next-generation deep-sequencing technologies [117].
Diagnosis and prognosis
miRNAs found in EVs may be an indicator of diagnosis and prognosis. Many researchers have shown that the miRNA profiles in EVs are significantly different in patients with high-metastatic tumors. The miRNAs that may potentially be used as diagnostic and prognostic markers are summarized in [116]. Cancer-specific markers of EVs have been identified for application to cancer diagnostics at early stages. Microarray analyses revealed the accumulation of let-7a, miR-1229, miR-1246, miR-150, miR-21, miR-223, and miR-23a in the EV-concentrated fraction from patients with colorectal cancer [118]. In prostate cancer, miR-141 and miR-375 are promising markers of metastasis in the blood circulation. In particular, miR-141 levels were shown to be 46-fold higher in metastatic patients than healthy individuals in plasma [22, 119]. Both miRNAs were significantly increased in EVs in sera from men with metastatic disease [120] (reviewed in [121, 122]). EVs derived from patients with ovarian cancer were found to present a diagnostic profile of 8 miRNAs (miR-21, miR-141, miR-200a, miR-200c, miR-200b, miR-203, miR-205 and miR-214) [91]. In EVs from lung adenocarcinoma patients, the profiles of 12 miRNAs (miR-17-3p, miR-21, miR-106a, miR-146, miR-155, miR-191, miR-192, miR-203, miR-205, miR-210, miR-212, and miR-214), which are diagnostic in lung tumor biopsies, showed similar pattern [123]. The profiling of miRNAs in EVs is expected as a powerful strategy for cancer diagnosis and prognosis.
One bottleneck in the detection of miRNAs in EVs in the arena of diagnostics is the methods used for the collection of EVs. Collecting EVs using ultracentrifugation is a conventional way to collect EVs, but it is time consuming. Collecting EVs through filtration and affinity purification based on surface proteins are possible alternative strategies [124]. For expecting clinical use, EV collection using microfluidic immunoaffinity method that can directly bring into transcriptome analysis has been developed [125].
Drug delivery systems
Extracellular vesicles are anticipated to be useful as a drug delivery system to deliver anti-tumor siRNAs, as nucleic acid medicines, specifically to cancer cells as a novel cancer therapy [126]. Although the native targeting system is not known, Ohno et al. engineered the surface of EVs to express the GE11 peptide or the EGF peptide, which is specifically recognized by EGFR. The engineered EVs were successfully delivered to EGFR-expressing breast cancer cells, and the encapsulated siRNAs repressed target gene expression [127]. Alvarez-Erviti et al. [128] also modified the surface of the exosome to deliver siRNA to brain in mice using self-derived exosomes from dendritic cells. For neuron-specific targeting, Lamp2b, and exosomal membrane protein, was fused to neuron-specific RVG peptide.
In addition, introducing exogenous siRNAs into exosome is another important aspect for drug delivery system. Wahlgren et al. [129] tested several methods to introduce double-stranded siRNA into plasma-derived exosome by electroporation and by chemical transfections using lipid micelles. They showed that lipid micelles themself transfer siRNA into cells and hard to distinguish from the delivery by exosome. Shtam et al. [130] also used both lipofectamine and electroporation to introduce siRNA into human exosomes. And they supported that electroporation is applicable for siRNA introduction into exosomes. Alvarez-Erviti et al. [128] used electroporation to introduce siRNA into exosomes. However, the efficiency of loading siRNAs by electroporation in exosome should be improved for drug delivery system, as the current efficiency is lower than 30 % [129].
Together with as-yet-undeveloped technologies for the targeted packaging of miRNAs, the targeting of EVs would be a useful technology in developing drug delivery systems for nucleic acid medicines.
Therapeutics
As miRNAs transferred by cancer-derived EVs are known to promote cancer progression in many ways, the removal of cancer-secreted EVs is assumed to represent an effective means of repressing cancer proliferation and dissemination. Aethlon Medical Inc. has already developed a dialysis-like system that removes EVs in the circulating blood via affinity binding to immobilized EV-binding lectins and antibodies [131]. Although some concerns should be addressed before this system is used for treatment, such as how much impact the transient reduction of cancer-derived EVs has and whether the removal of EVs not only from cancer cells but also from non-cancerous cells in the body has a negative impact, this method is anticipated to serve as a novel cancer therapy.
Conclusions and perspectives
MicroRNAs regulate gene expression levels post-transcriptionally. Not only do miRNAs function in the miRNA-coding cells, but they also appear to be functional in the recipient cells. miRNAs are secreted in a form protected by proteins or lipid bilayer vesicles, referred to as EVs. Increasing numbers of studies on EVs have revealed that EVs are an important cell–cell communication tool in biological activities and tumor progression.
EVs contain several molecules within one vesicle, carrying not only a single type of miRNA, but various types of miRNAs, mRNAs, proteins, and growth factors. The ability of EVs to transfer many types of molecules at the same time is a key feature of these vesicles compared with protein-bound miRNAs or other secreted biomolecules. In addition, cells are likely to take up multiple EVs simultaneously. Therefore, various types of molecules that they carry could have co-operative effects on the recipient cells. However, only a fraction of the miRNAs and molecules responsible for certain activities in the cells are known. To understand the true biological function of the roles of EVs, it will be necessary to evaluate the comprehensive effects of EVs on cells using different approaches.
miRNAs transferred by EVs function in tumor progression. Reducing cancer-derived EV uptake by inhibiting secretion and eliminating EVs would provide supportive therapeutic strategies to inhibit cancer progression and metastasis. In addition, enhancing or mimicking cancer-suppressive EVs could also be employed as a novel therapy.
References
Bartel D (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5):843–854. doi:10.1016/0092-8674(93)90529-Y
Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E (2005) Phylogenetic shadowing and computational identification of human microRNA genes. Cell 120(1):21–24. doi:10.1016/j.cell.2004.12.031
Gaidatzis D, van Nimwegen E, Hausser J, Zavolan M (2007) Inference of miRNA targets using evolutionary conservation and pathway analysis. BMC Bioinform 8:69. doi:10.1186/1471-2105-8-69
Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34(Database issue):D140–D144. doi:10.1093/nar/gkj112
van der Krol AR, Mur LA, Beld M, Mol JN, Stuitje AR (1990) Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 2(4):291–299. doi:10.1105/tpc.2.4.291
Napoli C, Lemieux C, Jorgensen R (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2(4):279–289. doi:10.1105/tpc.2.4.279
Romano N, Macino G (1992) Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol Microbiol 6(22):3343–3353. doi:10.1111/j.1365-2958.1992.tb02202.x
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811. doi:10.1038/35888
Montgomery MK, Xu S, Fire A (1998) RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc Natl Acad Sci USA 95:15502–15507. doi:10.1073/pnas.95.26.15502
Kim VN (2005) MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 6(5):376–385. doi:10.1038/nrm1644
Lai EC (2002) Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet 30(4):363–364. doi:10.1038/ng865
Lee I, Ajay SS, Yook JI, Kim HS, Hong SH, Kim NH, Dhanasekaran SM, Chinnaiyan AM, Athey BD (2009) New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res 19(7):1175–1183. doi:10.1101/gr.089367.108
Stroynowska-Czerwinska A, Fiszer A, Krzyzosiak WJ (2014) The panorama of miRNA-mediated mechanisms in mammalian cells. Cell Mol Life Sci 71(12):2253–2270. doi:10.1007/s00018-013-1551-6
Yoo AS, Staahl BT, Chen L, Crabtree GR (2009) MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460(7255):642–646. doi:10.1038/nature08139
Benetti R, Gonzalo S, Jaco I, Munoz P, Gonzalez S, Schoeftner S, Murchison E, Andl T, Chen T, Klatt P, Li E, Serrano M, Millar S, Hannon G, Blasco MA (2008) A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol 15(3):268–279. doi:10.1038/nsmb.1399
Jansson MD, Lund AH (2012) MicroRNA and cancer. Mol Oncol 6(6):590–610. doi:10.1016/j.molonc.2012.09.006
Iorio MV, Croce CM (2012) Causes and consequences of microRNA dysregulation. Cancer J 18(3):215–222. doi:10.1097/PPO.0b013e318250c001
Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K (2010) The microRNA spectrum in 12 body fluids. Clin Chem 56(11):1733–1741. doi:10.1373/clinchem.2010.147405
Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee M-LT, Schmittgen TD, Nana-Sinkam SP, Jarjoura D, Marsh CB (2008) Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE 3(11):e3694–e3694. doi:10.1371/journal.pone.0003694
Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N, Bentwich Z, Hod M, Goren Y, Chajut A (2008) Serum microRNAs are promising novel biomarkers. PLoS ONE 3(9):e3148–e3148. doi:10.1371/journal.pone.0003148
Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 105(30):10513–10518. doi:10.1073/pnas.0804549105
Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9:654–659. doi:10.1038/ncb1596
Noerholm M, Balaj L, Limperg T, Salehi A, Zhu LD, Hochberg FH, Breakefield XO, Carter BS, Skog J (2012) RNA expression patterns in serum microvesicles from patients with glioblastoma multiforme and controls. BMC Cancer 12:22. doi:10.1186/1471-2407-12-22
Hanke M, Hoefig K, Merz H, Feller AC, Kausch I, Jocham D, Warnecke JM, Sczakiel G (2010) A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol Oncol 28(6):655–661. doi:10.1016/j.urolonc.2009.01.027
Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, Abemayor E, Wong DT (2009) Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res 15(17):5473–5477. doi:10.1158/1078-0432.CCR-09-0736
Zubakov D, Boersma AW, Choi Y, van Kuijk PF, Wiemer EA, Kayser M (2010) MicroRNA markers for forensic body fluid identification obtained from microarray screening and quantitative RT-PCR confirmation. Int J Legal Med 124(3):217–226. doi:10.1007/s00414-009-0402-3
Kosaka N, Izumi H, Sekine K, Ochiya T (2010) MicroRNA as a new immune-regulatory agent in breast milk. Silence 1(1):7. doi:10.1186/1758-907X-1-7
Zhou Q, Li M, Wang X, Li Q, Wang T, Zhu Q, Zhou X, Wang X, Gao X, Li X (2012) Immune-related microRNAs are abundant in breast milk exosomes. Int J Biol Sci 8(1):118–123. doi:10.7150/ijbs.8.118
Blank A, Dekker CA (1981) Ribonucleases of human serum, urine, cerebrospinal fluid, and leukocytes. Activity staining following electrophoresis in sodium dodecyl sulfate–polyacrylamide gels. Biochemistry 20(8):2261–2267. doi:10.1021/bi00511a030
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY (2008) Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18(10):997–1006. doi:10.1038/cr.2008.282
Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M (2011) Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA 108(12):5003–5008. doi:10.1073/pnas.1019055108
Turchinovich A, Burwinkel B (2012) Distinct Ago1 and Ago2 associated miRNA profiles in human cells and blood plasma. RNA Biol 9(8):1066–1075. doi:10.4161/rna.21083
Turchinovich A, Weiz L, Langheinz A, Burwinkel B (2011) Characterization of extracellular circulating microRNA. Nucleic Acids Res 39(16):7223–7233. doi:10.1093/nar/gkr254
Wang K, Zhang S, Weber J, Baxter D, Galas DJ (2010) Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res 38(20):7248–7259. doi:10.1093/nar/gkq601
Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT (2011) MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 13(4):423–433. doi:10.1038/ncb2210
Gould SJ, Raposo G (2013) As we wait: coping with an imperfect nomenclature for extracellular vesicles. J Extracell Vesicles. doi:10.3402/jev.v2i0.20389
Simpson RJ, Mathivanan S (2012) Extracellular microvesicles: the need for internationally recognised nomenclature and stringent purification criteria. J Proteom Bioinform 05(02):10000. doi:10.4172/jpb.10000e10
Raposo G, Stoorvogel W (2013) Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200(4):373–383. doi:10.1083/jcb.201211138
Elmore S (2007) Apoptosis: a review of programmed cell death. Toxicol Pathol 35(4):495–516. doi:10.1080/01926230701320337
Cocucci E, Racchetti G, Meldolesi J (2009) Shedding microvesicles: artefacts no more. Trends Cell Biol 19(2):43–51. doi:10.1016/j.tcb.2008.11.003
Crescitelli R, Lasser C, Szabo TG, Kittel A, Eldh M, Dianzani I, Buzas EI, Lotvall J (2013) Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J Extracell Vesicles. doi:10.3402/jev.v2i0.20677
Akers JC, Gonda D, Kim R, Carter BS, Chen CC (2013) Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J Neurooncol 113(1):1–11. doi:10.1007/s11060-013-1084-8
Lai CP, Breakefield XO (2012) Role of exosomes/microvesicles in the nervous system and use in emerging therapies. Front Physiol 3:228. doi:10.3389/fphys.2012.00228
Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M (2008) Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319(5867):1244–1247. doi:10.1126/science.1153124
Bobrie A, Colombo M, Raposo G, Thery C (2011) Exosome secretion: molecular mechanisms and roles in immune responses. Traffic 12(12):1659–1668. doi:10.1111/j.1600-0854.2011.01225.x
Harding C, Heuser J, Stahl P (1983) Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol 97(2):329–339. doi:10.1083/jcb.97.2.329
Pan BT, Johnstone RM (1983) Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell 33(3):967–978. doi:10.1016/0092-8674(83)90040-5
Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C (1987) Vesicle formation during reticulocyte maturation. association of plasma membrane activities with released vesicles (exosomes). J Biol Chem 262(19):9412–9420
Trams E, Lauter C, Normansalem J, Heine U (1981) Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim Biophys Acta 645(1):63–70. doi:10.1016/0005-2736(81)90512-5
Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91(4):457–466. doi:10.1016/S0092-8674(00)80432-8
Kosaka N, Yoshioka Y, Hagiwara K, Tominaga N, Katsuda T, Ochiya T (2013) Trash or treasure: extracellular microRNAs and cell-to-cell communication. Front Genet 4:173. doi:10.3389/fgene.2013.00173
Hu G, Drescher KM, Chen XM (2012) Exosomal miRNAs: biological properties and therapeutic potential. Front Genet 3:56. doi:10.3389/fgene.2012.00056
Théry C (2011) Exosomes: secreted vesicles and intercellular communications. F1000 Prime Rep 3(15). doi:10.3410/B3-15
Katzmann DJ, Babst M, Emr SD (2001) Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106(2):145–155. doi:10.1016/S0092-8674(01)00434-2
Kowal J, Tkach M, Thery C (2014) Biogenesis and secretion of exosomes. Curr Opin Cell Biol 29C:116–125. doi:10.1016/j.ceb.2014.05.004
Stuffers S, Sem Wegner C, Stenmark H, Brech A (2009) Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 10(7):925–937. doi:10.1111/j.1600-0854.2009.00920.x
Andreu Z, Yanez-Mo M (2014) Tetraspanins in extracellular vesicle formation and function. Front Immunol 5:442. doi:10.3389/fimmu.2014.00442
Kosaka N, Iguchi H, Hagiwara K, Yoshioka Y, Takeshita F, Ochiya T (2013) Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem 288:10849–10859. doi:10.1074/jbc.M112.446831
Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T (2010) Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem 285(23):17442–17452. doi:10.1074/jbc.M110.107821
Chairoungdua A, Smith DL, Pochard P, Hull M, Caplan MJ (2010) Exosome release of β-catenin: a novel mechanism that antagonizes Wnt signaling. J Cell Biol 190(6):1079–1091. doi:10.1083/jcb.201002049
Tejera E, Rocha-Perugini V, Lopez-Martin S, Perez-Hernandez D, Bachir AI, Horwitz AR, Vazquez J, Sanchez-Madrid F, Yanez-Mo M (2013) CD81 regulates cell migration through its association with Rac GTPase. Mol Biol Cell 24(3):261–273. doi:10.1091/mbc.E12-09-0642
van Niel G, Charrin S, Simoes S, Romao M, Rochin L, Saftig P, Marks MS, Rubinstein E, Raposo G (2011) The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev Cell 21(4):708–721. doi:10.1016/j.devcel.2011.08.019
Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, Bond VC, Borras FE, Breakefield X, Budnik V, Buzas E, Camussi G, Clayton A, Cocucci E, Falcon-Perez JM, Gabrielsson S, Gho YS, Gupta D, Harsha HC, Hendrix A, Hill AF, Inal JM, Jenster G, Kramer-Albers EM, Lim SK, Llorente A, Lotvall J, Marcilla A, Mincheva-Nilsson L, Nazarenko I, Nieuwland R, Nolte-’t Hoen EN, Pandey A, Patel T, Piper MG, Pluchino S, Prasad TS, Rajendran L, Raposo G, Record M, Reid GE, Sanchez-Madrid F, Schiffelers RM, Siljander P, Stensballe A, Stoorvogel W, Taylor D, Thery C, Valadi H, van Balkom BW, Vazquez J, Vidal M, Wauben MH, Yanez-Mo M, Zoeller M, Mathivanan S (2012) Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol 10(12):e1001450. doi:10.1371/journal.pbio.1001450
Kim DK, Kang B, Kim OY, Choi DS, Lee J, Kim SR, Go G, Yoon YJ, Kim JH, Jang SC, Park KS, Choi EJ, Kim KP, Desiderio DM, Kim YK, Lotvall J, Hwang D, Gho YS (2013) EVpedia: an integrated database of high-throughput data for systemic analyses of extracellular vesicles. J Extracell Vesicles. doi:10.3402/jev.v2i0.20384
Choi DS, Kim DK, Kim YK, Gho YS (2013) Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics 13(10–11):1554–1571. doi:10.1002/pmic.201200329
Choi DS, Kim DK, Kim YK, Gho YS (2014) Proteomics of extracellular vesicles: exosomes and ectosomes. Mass Spectrom Rev. doi:10.1002/mas.21420
Jenjaroenpun P, Kremenska Y, Nair VM, Kremenskoy M, Joseph B, Kurochkin IV (2013) Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing. PeerJ 1:e201. doi:10.7717/peerj.201
Kruger S, Abd Elmageed ZY, Hawke DH, Worner PM, Jansen DA, Abdel-Mageed AB, Alt EU, Izadpanah R (2014) Molecular characterization of exosome-like vesicles from breast cancer cells. BMC Cancer 14:44. doi:10.1186/1471-2407-14-44
Wolfers J, Lozier A, Raposo G, Regnault A, Thery C, Masurier C, Flament C, Pouzieux S, Faure F, Tursz T, Angevin E, Amigorena S, Zitvogel L (2001) Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat Med 7(3):297–303. doi:10.1038/85438
Aalberts M, van Dissel-Emiliani FM, van Adrichem NP, van Wijnen M, Wauben MH, Stout TA, Stoorvogel W (2012) Identification of distinct populations of prostasomes that differentially express prostate stem cell antigen, annexin A1, and GLIPR2 in humans. Biol Reprod 86(3):82. doi:10.1095/biolreprod.111.095760
Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, Bernad A, Sanchez-Madrid F (2011) Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun 2:282. doi:10.1038/ncomms1285
Pigati L, Yaddanapudi SC, Iyengar R, Kim DJ, Hearn SA, Danforth D, Hastings ML, Duelli DM (2010) Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS ONE 5(10):e13515. doi:10.1371/journal.pone.0013515
Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sanchez-Madrid F (2013) Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun 4:2980. doi:10.1038/ncomms3980
Feng D, Zhao WL, Ye YY, Bai XC, Liu RQ, Chang LF, Zhou Q, Sui SF (2010) Cellular internalization of exosomes occurs through phagocytosis. Traffic 11(5):675–687. doi:10.1111/j.1600-0854.2010.01041.x
Fitzner D, Schnaars M, van Rossum D, Krishnamoorthy G, Dibaj P, Bakhti M, Regen T, Hanisch UK, Simons M (2011) Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J Cell Sci 124(Pt 3):447–458. doi:10.1242/jcs.074088
Tian T, Zhu YL, Zhou YY, Liang GF, Wang YY, Hu FH, Xiao ZD (2014) Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J Biol Chem 289(32):22258–22267. doi:10.1074/jbc.M114.588046
Parolini I, Federici C, Raggi C, Lugini L, Palleschi S, De Milito A, Coscia C, Iessi E, Logozzi M, Molinari A, Colone M, Tatti M, Sargiacomo M, Fais S (2009) Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem 284(49):34211–34222. doi:10.1074/jbc.M109.041152
Escrevente C, Keller S, Altevogt P, Costa J (2011) Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer 11:108. doi:10.1186/1471-2407-11-108
Mulcahy LA, Pink RC, Carter DR (2014) Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. doi:10.3402/jev.v3.24641
Morelli AE, Larregina AT, Shufesky WJ, Sullivan ML, Stolz DB, Papworth GD, Zahorchak AF, Logar AJ, Wang Z, Watkins SC, Falo LD Jr, Thomson AW (2004) Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood 104(10):3257–3266. doi:10.1182/blood-2004-03-0824
Christianson HC, Svensson KJ, van Kuppevelt TH, Li JP, Belting M (2013) Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc Natl Acad Sci USA 110(43):17380–17385. doi:10.1073/pnas.1304266110
Svensson KJ, Christianson HC, Wittrup A, Bourseau-Guilmain E, Lindqvist E, Svensson LM, Morgelin M, Belting M (2013) Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid raft-mediated endocytosis negatively regulated by caveolin-1. J Biol Chem 288(24):17713–17724. doi:10.1074/jbc.M112.445403
Tian T, Wang Y, Wang H, Zhu Z, Xiao Z (2010) Visualizing of the cellular uptake and intracellular trafficking of exosomes by live-cell microscopy. J Cell Biochem 111(2):488–496. doi:10.1002/jcb.22733
Franzen CA, Simms PE, Van Huis AF, Foreman KE, Kuo PC, Gupta GN (2014) Characterization of uptake and internalization of exosomes by bladder cancer cells. Biomed Res Int 2014:619829. doi:10.1155/2014/619829
Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Wurdinger T, Middeldorp JM (2010) Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA 107(14):6328–6333. doi:10.1073/pnas.0914843107
Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, Sun F, Lu J, Yin Y, Cai X, Sun Q, Wang K, Ba Y, Wang Q, Wang D, Yang J, Liu P, Xu T, Yan Q, Zhang J, Zen K, Zhang CY (2010) Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell 39(1):133–144. doi:10.1016/j.molcel.2010.06.010
Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan MLG, Karlsson JM, Baty CJ, Gibson GA, Erdos G, Wang ZL, Milosevic J, Tkacheva OA, Divito SJ, Jordan R, Lyons-Weiler J, Watkins SC, Morelli AE (2012) Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 119(3):756–766. doi:10.1182/blood-2011-02-338004
Fruhbeis C, Frohlich D, Kuo WP, Amphornrat J, Thilemann S, Saab AS, Kirchhoff F, Mobius W, Goebbels S, Nave KA, Schneider A, Simons M, Klugmann M, Trotter J, Kramer-Albers EM (2013) Neurotransmitter-triggered transfer of exosomes mediates oligodendrocyte–neuron communication. PLoS Biol 11(7):e1001604. doi:10.1371/journal.pbio.1001604
Tickner JA, Urquhart AJ, Stephenson SA, Richard DJ, O’Byrne KJ (2014) Functions and therapeutic roles of exosomes in cancer. Front Oncol 4:127. doi:10.3389/fonc.2014.00127
Taylor DD, Gercel-Taylor C (2008) MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 110(1):13–21. doi:10.1016/j.ygyno.2008.04.033
Rosell R, Wei J, Taron M (2009) Circulating microRNA signatures of tumor-derived exosomes for early diagnosis of non-small-cell lung cancer. Clin Lung Cancer 10(1):8–9. doi:10.3816/CLC.2009.n.001
Bobrie A, Krumeich S, Reyal F, Recchi C, Moita LF, Seabra MC, Ostrowski M, Thery C (2012) Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res 72(19):4920–4930. doi:10.1158/0008-5472.CAN-12-0925
Roma-Rodrigues C, Fernandes AR, Baptista PV (2014) Exosome in tumour microenvironment: overview of the crosstalk between normal and cancer cells. Biomed Res Int 2014:179486. doi:10.1155/2014/179486
Qin Q, Furong W, Baosheng L (2014) Multiple functions of hypoxia-regulated miR-210 in cancer. J Exp Clin Cancer Res 33:50. doi:10.1186/1756-9966-33-50
Zhuang G, Wu X, Jiang Z, Kasman I, Yao J, Guan Y, Oeh J, Modrusan Z, Bais C, Sampath D, Ferrara N (2012) Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J 31(17):3513–3523. doi:10.1038/emboj.2012.183
Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, Yu Y, Chow A, O’Connor ST, Chin AR, Yen Y, Wang Y, Marcusson EG, Chu P, Wu J, Wu X, Li AX, Li Z, Gao H, Ren X, Boldin MP, Lin PC, Wang SE (2014) Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 25(4):501–515. doi:10.1016/j.ccr.2014.03.007
Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky AM, Breakefield XO (2008) Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 10(12):1470–1476. doi:10.1038/ncb1800
Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T (2007) MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133(2):647–658. doi:10.1053/j.gastro.2007.05.022
Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H (2008) MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 27(15):2128–2136. doi:10.1038/sj.onc.1210856
Rupp AK, Rupp C, Keller S, Brase JC, Ehehalt R, Fogel M, Moldenhauer G, Marme F, Sultmann H, Altevogt P (2011) Loss of EpCAM expression in breast cancer derived serum exosomes: role of proteolytic cleavage. Gynecol Oncol 122(2):437–446. doi:10.1016/j.ygyno.2011.04.035
Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T (2004) Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 64(11):3753–3756. doi:10.1158/0008-5472.CAN-04-0637
Ohshima K, Inoue K, Fujiwara A, Hatakeyama K, Kanto K, Watanabe Y, Muramatsu K, Fukuda Y, Ogura S, Yamaguchi K, Mochizuki T (2010) Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS ONE 5(10):e13247. doi:10.1371/journal.pone.0013247
Karrison TG, Ferguson DJ, Meier P (1999) Dormancy of mammary carcinoma after mastectomy. J Natl Cancer Inst 91(1):80–85. doi:10.1093/jnci/91.1.80
Ono M, Kosaka N, Tominaga N, Yoshioka Y, Takeshita F, Takahashi RU, Yoshida M, Tsuda H, Tamura K, Ochiya T (2014) Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Sci Signal 7(332):63. doi:10.1126/scisignal.2005231
Chen WX, Liu XM, Lv MM, Chen L, Zhao JH, Zhong SL, Ji MH, Hu Q, Luo Z, Wu JZ, Tang JH (2014) Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PLoS ONE 9(4):e95240. doi:10.1371/journal.pone.0095240
Zhang HG, Grizzle WE (2011) Exosomes and cancer: a newly described pathway of immune suppression. Clin Cancer Res 17(5):959–964. doi:10.1158/1078-0432.CCR-10-1489
Clayton A (2012) Cancer cells use exosomes as tools to manipulate immunity and the microenvironment. Oncoimmunology 1(1):78–80. doi:10.4161/onci.1.1.17826
Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, Zanesi N, Crawford M, Ozer GH, Wernicke D, Alder H, Caligiuri MA, Nana-Sinkam P, Perrotti D, Croce CM (2012) MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA 109(31):E2110–E2116. doi:10.1073/pnas.1209414109
Yang M, Chen J, Su F, Yu B, Su F, Lin L, Liu Y, Huang JD, Song E (2011) Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer 10:117. doi:10.1186/1476-4598-10-117
Alderton GK (2012) Metastasis. Exosomes drive premetastatic niche formation. Nat Rev Cancer 12(7):447. doi:10.1038/nrc3304
Grange C, Tapparo M, Collino F, Vitillo L, Damasco C, Deregibus MC, Tetta C, Bussolati B, Camussi G (2011) Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res 71(15):5346–5356. doi:10.1158/0008-5472.CAN-11-0241
Rana S, Malinowska K, Zoller M (2013) Exosomal tumor microRNA modulates premetastatic organ cells. Neoplasia 15(3):281–295. doi:10.1593/neo.122010
Kosaka N, Iguchi H, Yoshioka Y, Hagiwara K, Takeshita F, Ochiya T (2012) Competitive interactions of cancer cells and normal cells via secretory microRNAs. J Biol Chem 287(2):1397–1405. doi:10.1074/jbc.M111.288662
Gastpar R, Gehrmann M, Bausero MA, Asea A, Gross C, Schroeder JA, Multhoff G (2005) Heat shock protein 70 surface-positive tumor exosomes stimulate migratory and cytolytic activity of natural killer cells. Cancer Res 65(12):5238–5247. doi:10.1158/0008-5472.CAN-04-3804
Kosaka N, Iguchi H, Ochiya T (2010) Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 101:2087–2092. doi:10.1111/j.1349-7006.2010.01650.x
Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, Liang M, Dittmar RL, Liu Y, Liang M, Kohli M, Thibodeau SN, Boardman L, Wang L (2013) Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom 14:319. doi:10.1186/1471-2164-14-319
Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H, Watanabe M, Nakagama H, Yokota J, Kohno T, Tsuchiya N (2014) Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 9(4):e92921. doi:10.1371/journal.pone.0092921
Yaman Agaoglu F, Kovancilar M, Dizdar Y, Darendeliler E, Holdenrieder S, Dalay N, Gezer U (2011) Investigation of miR-21, miR-141, and miR-221 in blood circulation of patients with prostate cancer. Tumour Biol 32(3):583–588. doi:10.1007/s13277-011-0154-9
Hessvik NP, Phuyal S, Brech A, Sandvig K, Llorente A (2012) Profiling of microRNAs in exosomes released from PC-3 prostate cancer cells. Biochim Biophys Acta 1819(11–12):1154–1163. doi:10.1016/j.bbagrm.2012.08.016
Hessels D, Schalken JA (2013) Urinary biomarkers for prostate cancer: a review. Asian J Androl 15(3):333–339. doi:10.1038/aja.2013.6
Hessvik NP, Sandvig K, Llorente A (2013) Exosomal miRNAs as biomarkers for prostate cancer. Front Genet 4:36. doi:10.3389/fgene.2013.00036
Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH (2009) Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer 10(1):42–46. doi:10.3816/CLC.2009.n.006
Taylor DD, Zacharias W, Gercel-Taylor C (2011) Exosome isolation for proteomic analyses and RNA profiling. Methods Mol Biol 728:235–246. doi:10.1007/978-1-61779-068-3_15
Chen C, Skog J, Hsu CH, Lessard RT, Balaj L, Wurdinger T, Carter BS, Breakefield XO, Toner M, Irimia D (2010) Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab Chip 10(4):505–511. doi:10.1039/b916199f
Kosaka N, Takeshita F, Yoshioka Y, Hagiwara K, Katsuda T, Ono M, Ochiya T (2013) Exosomal tumor-suppressive microRNAs as novel cancer therapy: “exocure” is another choice for cancer treatment. Adv Drug Deliv Rev 65(3):376–382. doi:10.1016/j.addr.2012.07.011
Ohno S, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, Fujita K, Mizutani T, Ohgi T, Ochiya T, Gotoh N, Kuroda M (2013) Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther 21(1):185–191. doi:10.1038/mt.2012.180
Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ (2011) Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol 29(4):341–345. doi:10.1038/nbt.1807
Wahlgren J, De L Karlson T, Brisslert M, Vaziri Sani F, Telemo E, Sunnerhagen P, Valadi H (2012) Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res 40(17):e130. doi:10.1093/nar/gks463
Shtam TA, Kovalev RA, Varfolomeeva EY, Makarov EM, Kil YV, Filatov MV (2013) Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Commun Signal CCS 11:88. doi:10.1186/1478-811X-11-88
Marleau AM, Chen CS, Joyce JA, Tullis RH (2012) Exosome removal as a therapeutic adjuvant in cancer. J Transl Med 10:134. doi:10.1186/1479-5876-10-134
Acknowledgments
This work was supported in part by a Grant-in-Aid for the Third-Term Comprehensive 10-Year Strategy for Cancer Control from the Ministry of Health, Labor, and Welfare, Japan; the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation; the New Energy and Industrial Technology Development Organization (NEDO); the Japan Society for the Promotion of Science through the Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Program) initiated by the Council for Science and Technology Policy; and a Grant-in-Aid for JSPS Fellows (No. 269202). The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Nishida-Aoki, N., Ochiya, T. Interactions between cancer cells and normal cells via miRNAs in extracellular vesicles. Cell. Mol. Life Sci. 72, 1849–1861 (2015). https://doi.org/10.1007/s00018-014-1811-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-014-1811-0