Abstract
Tbx3, a T-box transcription factor, regulates key steps in development of the heart and other organ systems. Here, we identify Sox4 as an interacting partner of Tbx3. Pull-down and nuclear retention assays verify this interaction and in situ hybridization reveals Tbx3 and Sox4 to co-localize extensively in the embryo including the atrioventricular and outflow tract cushion mesenchyme and a small area of interventricular myocardium. Tbx3, SOX4, and SOX2 ChIP data, identify a region in intron 1 of Gja1 bound by all tree proteins and subsequent ChIP experiments verify that this sequence is bound, in vivo, in the developing heart. In a luciferase reporter assay, this element displays a synergistic antagonistic response to co-transfection of Tbx3 and Sox4 and in vivo, in zebrafish, drives expression of a reporter in the heart, confirming its function as a cardiac enhancer. Mechanistically, we postulate that Sox4 is a mediator of Tbx3 transcriptional activity.
Similar content being viewed by others
Introduction
The T-box genes encode a phylogenetically conserved family of transcription factors that share a common DNA-binding motif known as the T-box domain. They play crucial roles in development where they are implicated in patterning, early cell-fate decisions, and many aspects of organogenesis [1]. Mutations of T-box genes have been associated with human disorders such as DiGeorge, Holt-Oram, and ulnar-mammary syndromes [2, 3].
Tbx2 and Tbx3 are closely related homologues of the T-box family that are expressed in many overlapping areas during development, including the heart, limbs, and lungs [1]. They typically function as transcriptional repressors and have been shown to have many, if not all, target genes in common, including regulators of the cell cycle [1]. In addition to their roles during development, Tbx2 and Tbx3 are also found over-expressed in melanoma, breast, and pancreatic cancers [4–6]. Their role in cancer may be related to their capacity to bypass senescence by repressing expression of p14ARF and P21CIP1 [7–9].
During heart development, Tbx3 is required for development of the cardiac conduction system and outflow tract [10–13]. In the myocardium of the sinus node and the atrioventricular bundle, Tbx3 represses a chamber myocardium-specific gene program, including the gap junction genes Gja1 and Gja5, encoding connexin 43 (Cx43) and Cx40 respectively, and natriuretic peptide precursor type A (Nppa). The hypothesis has thus been put forward that Tbx3 functions by imposing a primitive ‘nodal’ like phenotype on this early myocardium [10, 14–16]. Furthermore, Tbx3 null mice display defects in outflow tract development that have implied a role of Tbx3 in cardiac neural crest development and signaling between neural crest and the second heart field [11, 12]. Although these results have provided valuable insights into the roles of Tbx3 during multiple aspects of heart development, many of the underlying molecular mechanisms remain to be elucidated.
There are basically two aspects that dictate transcription factor binding to promoters and regulatory gene elements; the DNA sequence that is recognized and bound by the transcription factor and its repertoire of specific protein–protein interactions that can be made with other regulatory proteins. Both of these elements will define the ultimate transcriptional function of the factor and hence its downstream gene targets. The T-box factors Tbx2, Tbx3 and Tbx5 are known, for instance, to bind the homeobox protein Nkx2.5 [13, 17–19]. Since T-box factors are expressed and are required for the development of many different organs and tissues, complex forming with a factor such as Nkx2.5, which has a more cardiac-restricted expression pattern, may be instrumental in determining a set of heart specific T-box target genes. With the recent advent of ChIP-seq [20] a physical map of a transcription factor’s genome-wide DNA-binding profile can be generated. Combining datasets generated from different transcription factors, especially those known to interact, to search for small overlapping regions of binding, can be a powerful technique in defining regulatory elements, such as enhancers, and co-regulated genes.
While insights into the protein–protein interactions of a transcription factor provides useful molecular information, defining the function of the interaction in vivo, particularly in higher eukaryotes, can be a long and challenging path. Here, we describe a novel protein–protein interaction between Tbx3 and Sox4. Expression analysis shows multiple sites of coexpression in- and outside the embryonic heart at which this interaction may be functional. Their interaction was subsequently verified using both in vitro and sub-cellular localization assays. To explore the functional relevance of this novel interaction, heart-specific Tbx3 ChIP-seq data was compared to ChIP-seq and ChIP-chip data available for SOX2 and SOX4, which lead to the identification of a 1-kb regulatory element in intron 1 of Gja1 that is bound by both Tbx3, Sox4, and P300 in the developing mouse heart. In vitro, this element could activate a basal promoter and could be used to demonstrate a synergistic interaction between Sox4 and Tbx3. Its specific functionality as a cardiac enhancer could also be demonstrated, in vivo, using a zebrafish model system.
Materials and methods
Plasmid constructs
Full-length (aa 1–723/743) and T-box region (aa 94–300/320) of Tbx3 or Tbx3 isoform2 (+exon 2a) were PCR amplified from human cDNA (NM_005996/NM_016569) and cloned into pMAL2C (Clontech) to generate MBP fusion constructs. Full-length (aa 1–440) and N-terminal fragments (aa1–153, aa1–136, aa1–125) of SOX4 were PCR amplified from mouse cDNA (NM_009238) and cloned into pRP256nb to generate GST fusion constructs, or into pcDNA-myc (full length only) to generate myc-SOX4. Constructs encoding MBP-Tbx2-T-box, MBP-Tbx5-T-box, GST-Nkx2.5, HA-Tbx3, myc-Nkx2.5 have been described before [10, 21].
Yeast 2-hybrid screen
The T-box region of mouse Tbx3+2a (aa 94–320, NM_198052) was cloned into pGBKT7 (Clontech) and tested for self-activation by co-transfection to yeast strain AH109 (Clontech) with empty activation domain (AD) plasmid pGADT7 (Clontech). Bait construct was transformed into AH109, which was subsequently mated with yeast strain Y187 that was pretransformed with prey library of mouse embryonic day (E) 11.5 cDNA (Clontech) according to the manufacturer’s instructions. Clones were selected on triple-drop-out selection media lacking leucine, tryptophan and histidine in the presence of the galactoside X-α-Gal. Surviving colonies were replated to triple drop out medium and subsequently picked for AD-plasmid rescue and sequencing.
In vitro protein interactions assay
MBP pulldown assays were performed as described before [21], using anti-GST (GST-2, Sigma-Aldrich) as primary antibody for Western detection.
Immunofluorescence
Cells were transfected with 375 ng DNA of each plasmid, empty vector was added such that all cells received the same amount of total DNA. Primary antibodies used were rabbit anti-HA (H6908, Sigma-Aldrich), mouse anti-myc (9E10, Santa-Cruz) at 1:250 dilutions, and secondary antibodies were Alexa Fluor 488 goat anti-rabbit IgG and Alexa Fluor 568 goat anti-mouse IgG (Molecular Probes), at 1:250 dilutions. TO-PRO3 (Invitrogen) was used for nuclear counterstaining. Immunofluorescent detection of proteins was repeated at least three times, and representative examples were photographed on a Leica DM5500 confocal laser microscope (Leica).
In situ hybridization
In situ hybridization was performed as described before on 10-μm-thick sections [22]. T-box antisense probes have been described previously [23]. Sox4 probe was generated using a template based on the 3′UTR of Sox4 (1,885–2,886 of mouse Sox4 mRNA (NM_009238)).
ChIP data-analysis
Conditional Tbx3 over-expressing and cardiac specific tamoxifen inducible Cre (Mer-Cre-Mer) mice have been described before [10, 24]. Male mouse hearts were isolated 4 days after intra-peritoneal injections of tamoxifen, and Tbx3 over-expression was confirmed by qRT-PCR, in situ hybridization and immunohistochemistry (not shown). ChIP was performed on mouse hearts using anti-Tbx3 (A-20, Santa-Cruz). In this case Mer-Cre-Mer mice, lacking the Tbx3 expression construct, injected with tamoxifen served as ChIP control. Isolated DNA fragments were analyzed using high-throughput sequencing (data and analysis will be published elsewhere). Data significance of Tbx3-binding peaks were analyzed using a Fisher's exact test with comparison to ChIP control data. SOX4 and SOX2 ChIP data were obtained from NCBI gene expression omnibus (accession: GSE11874; [25, 26]) and analyses on data were carried using the Web-based software Galaxy (http://galaxy.psu.edu/). Annotated genes co-occurring in both assays were selected for further analysis.
Transcription factor binding site prediction
To identify potential Sox4 and Tbx3-binding sites, high-quality position weight matrices from the Jaspar database were used (http://jaspar.genereg.net/; MA0009.1 for T-box-binding sites; MA0077.1, MA0078.1, MA0084.1, MA0087.1, MA0143.1 and MA0442.1 for Sox HMG-box-binding sites). In addition, the predicted wwCAAwG sequence for Sox4 binding was searched [27]. Relative score threshold was set to 85% (Sox) or 70% (Tbx).
In vivo ChIP
For Tbx3 and SOX4 ChIP experiments, 36 hearts of ED10.5 wild-type mouse embryos were isolated and fixed at room temperature for 15 min with 1% formaldehyde. Cells were lysed and Dounce homogenized. Cross-linked nuclei were sonicated to obtain chromatin fragments with average size of ~400 bp. Pre-cleared chromatin fragments were incubated at 4°C for 4 h with 10 μg antibodies against Tbx3 (A-20, sc-17871, Santa Cruz Biotechnology) or SOX4 (C-20, sc-17326, Santa Cruz Biotechnology). Protein G beads were added to capture the chromatin-antibody complex. After five washing steps, the protein-DNA complex was eluted with 100 mM NaHCO3 and 1% SDS at room temperature, and cross-linking was reversed by incubating at 65°C overnight. After RNaseA and Proteinase K treatments, the DNA fragments were purified by phenol–chloroform, precipitated in ethanol and dissolved in 50 μl H2O, and analyzed using PCR. PCR primers are listed in Table 1. p300 ChIP was performed in wild-type adult mouse heart using an antibody against p300 (C-20, sc-585, Santa Cruz Biotechnology) as described above, with the modification of cross-linking for 1 h with 2% formaldehyde. In all cases, control PCRs represent regions found not to bind Tbx3 in ChIP-seq dataset or SOX4 based on the published SOX4 ChIP data.
ChIP QPCR reactions were performed and analyzed as described previously [21]. In vivo ChIP reactions were performed as described above, using 40 embryonic (ED 10.5) mouse hearts and matched IgG antibody (Santa Cruz, sc-2028) as a negative control.
Zebrafish enhancer assay
The putative Gja1 enhancer sequence was cloned into pGEM-T Easy (Promega) and amplified by PCR. The resulting PCR product was then cloned in a plasmid containing the e1b minimal fish promoter driving the expression of a H2A-eGFP fusion protein, upstream of the e1b sequence, generating pTOL2-EnhGja1-H2AeGFP.
pTOL2-EnhGja1-H2AeGFP was injected in zebrafish embryos at 1-cell stage at a final concentration of 10 ng/μl in presence of 25 ng/μl TOL2 transposase RNA. Embryos were subsequently kept at 28.5°C in E3 medium and imaged at 72 hpf.
Luciferase assays
COS7 cells, grown in 12-well plates in DMEM supplemented with 10% FCS (Gibco-BRL) and glutamine, were transfected using polyethylenimine 25 kDa (PEI) (Brunschwick) at a 1:3 ratio (DNA:PEI). Reporter construct was generated by ligating Cx43 putative enhancer region (chr10:56,097,392–56,098,369) to pGL2basic+minimal promoter (control reporter). Standard transfections used 1.6 μg of reporter (or control reporter) vector cotransfected with 3 ng phRG-TK Renilla vector (Promega) as normalization control. pCDNA3 constructs expressing Tbx2, Tbx3, Tbx5, and SOX4 were cotransfected as appropriate. Transfections were carried out at least four times and measured in duplo. Luciferase measurements were performed using a Promega Turner Biosystems Modulus Multimode Reader luminometer. All data was statistically validated using an ANOVA two-way test for all combinations of Sox4 and T-box.
Results
Tbx3 interacts with Sox4
To gain further insight into the molecular mechanisms by which Tbx3 controls gene expression, we performed a yeast 2-hybrid screen with Tbx3 as bait. From an initial screen of >1 × 106 colonies, 12 surviving clones revealed a GAL4 fusion to a peptide (>40 aa) in a reading frame coding for a BLASTP genome identifiable sequence. Two of these clones encoded an N-terminal fragment of Sox4, a high mobility group (HMG) domain containing transcription factor that has been previously shown to be essential for normal outflow tract development and atrioventricular valve formation [28–30]. The fragment encodes amino acids 3–153 of mouse Sox4, which contains the entire HMG domain. No other functional domains have been identified within this part of the protein and a database search for conserved domains using the NCBI CDD search option revealed no other conserved domains in this fragment (Fig. 1a; [31–33]).
The T-box of Tbx3 interacts with the HMG domain of SOX4. a Diagram showing full-length SOX4 with conserved domains, and the clone that was identified in our screen (SOX4-N). The sequence of a fragment containing the 3rd α-helix (underlined) of the HMG box (bold) and its C-terminal tail (italics) is shown, with the positions of truncated constructs (125, 136). b MBP pulldown assays showing that GST tagged Nkx2.5, SOX4 and SOX4-N bind to MBP-Tbx3 (middle panel) but not MBP alone (left). The T-box domain only of Tbx3 and that of Tbx2, Tbx3+2a and Tbx5 retain the ability to bind to the HMG domain of SOX4 (right). c Mapping of the interaction domain of SOX4 showing that the construct that misses the C-terminal tail (SOX4-125) does not interact with the T-box, whereas longer constructs do. CD Central domain, S serine-rich region, TA transactivation domain
Tbx3 and Sox4 interact via their DNA-binding domains
To validate and further investigate the interaction between Tbx3 and Sox4, we performed in vitro binding assays using bacterially expressed Tbx3 fused to MBP and Sox4 fused to GST. Both full-length Sox4 and the N-terminal fragment that was identified in the screen, are able to interact with MBP-Tbx3, but not with MBP alone (Fig. 1b). We next tested whether binding of Tbx3 to Sox4 is unique among T-box proteins, or whether the cardiac expressed T-box proteins Tbx2 and Tbx5 can also bind to Sox4. We found that the T-box of Tbx2 and Tbx5 are able to bind the N-terminal Sox4 fragment as well (Fig. 1b), suggesting a level of binding promiscuity between Sox4 and T-box proteins. Further, the apparently redundant isoform of Tbx3 (Tbx3+2a) [34], which differs from Tbx3 by a single 20 amino acid insertion within the T-box domain, shows similar binding properties (Fig. 1b).
Multiple bands were observed in the binding between full-length Sox4 and Tbx3 (Fig. 1b), which likely represent carboxy-terminal specific protein degradation by Escherichia coli endoproteases or premature GST-fused termination products. Strikingly, the size of the smallest of these products that still interacts with Tbx3 equals the size of the N-terminal fragment that was picked up in the two-hybrid screen. Smaller protein fragments, therefore, do not interact with Tbx3, indicating that further shortening of Sox4 would disrupt the interaction domain. To test this hypothesis, we compared binding of three N-terminal fragments (Fig. 1c). Stepwise truncation of Sox4 showed that the shortest construct, 136 residues in length that still binds Tbx3 contains the full HMG domain. Shortening this construct further to 125 residues results in a complete loss of interaction.
In summary, our in vitro binding assays show a strong interaction between Tbx3 and Sox4, which is mediated by their conserved DNA-binding regions; the T-box and the HMG-domain.
Tbx3 and Sox4 interact in a mammalian cellular context
To address whether the interaction between Tbx3 and Sox4 can also occur in mammalian cells, we analyzed the sub-cellular distribution of HA-tagged Tbx3 by immunofluorescence in HEK293 cells. When transfected to HEK cells, both Tbx3 isoforms are localized primarily in the cytoplasm, although some nuclear localization can be detected (Fig. 2) (Tbx3+2a data not shown). This behavior is unique for this cell line and is not observed in other cell lines such as COS7 or the cardiac H10 cell line (not shown) were Tbx3 is found almost exclusively in the nucleus. The cardiac transcription factor Nkx2.5, a known interaction partner of Tbx3, and Sox4 both localize to the nucleus when singularly transfected to HEK cells (Fig. 2). Upon co-transfection of Tbx3 with either Nkx2.5 or Sox4, Tbx3 could be detected nearly exclusively in the nucleus of HEK cells (Fig. 2), showing that both Nkx2.5 and Sox4 can interact with Tbx3 and facilitate its retention within the nucleus. The absence of nuclear retention of Tbx3 upon co-transfection of non-interacting nuclear localized GFP confirmed that the interaction was specific for Sox4 and Nkx2.5.
Tbx3 and SOX4 interact in HEK293 cells. Cells were transfected with expression constructs for HA-tagged Tbx3 in the presence or absence of nls-eYFP, SOX4, or Nkx2.5 (myc-tagged). Cytoplasmic Tbx3 is efficiently relocalized to the nucleus upon co-expression of SOX4 and Nkx2.5, whereas co-expression of the unrelated eYFP protein does not influence subcellular localization of Tbx3
Tbx3 and Sox4 are co-expressed during heart development
The observation that Tbx3 and Sox4 interact in vitro and in mammalian cells raises the question whether these proteins also interact during development. To determine in which tissues such a molecular interaction may occur, we compared the expression patterns of Sox4 and Tbx3 and the closely related Tbx2 and Tbx5 genes using in situ hybridization analysis of E11.5 mouse embryos. Sox4 is coexpressed with Tbx2, Tbx3 and Tbx5 in the thoracic body wall, mandibular component of the first branchial arch, the developing lungs, and the midgut (Fig. 3) [19, 23]. In the heart, Sox4 expression in the endocardium and mesenchyme of the cardiac cushions overlaps with Tbx2 and Tbx3. We also detect Sox4 expression in the ventral aspect of the interventricular ring, a subpopulation of primitive myocardium at the border of the left ventricle and outflow tract (Figs. 3, 4); [35].
Sagittal sections of E11.5 mouse embryos showing colocalization of Sox4 with T-box factors at multiple sites. a Consecutive sections of mouse embryo showing colocalization of Sox4 with Tbx2 and Tbx3 in mandibular component of the first branchial arch, and the midgut and with Tbx2, Tbx3 and Tbx5 in the developing heart, lungs and body wall. cTnI marks all myocardium. b Expression of Sox4 in the heart is localized in the endocardium and mesenchyme of the atrioventricular (*) outflow tract cushions (#), sites of abundant Tbx2 and Tbx3 expression. Tbx2 and Tbx3 are also expressed in the atrioventricular myocardium underlying the cushions, a region that does not express Sox4. Dotted lines mark contours of the myocardium. m Mandibular component, li liver, lu lung, th thyroid, mg midgut, bw body wall, a atrial lumen, v ventricular lumen, oft(m) outflow tract (myocardium), ift inflow tract, end endocardium, mes cushion mesenchyme, avc(m) atrioventricular canal (myocardium), sv sinus vinosus, cr cranial, ca caudal, ve ventral, do dorsal
Sox4 is expressed in the ventral aspect of the interventricular ring. In situ hybridization of E11.5 mouse heart showing cTnI stained myocardium, Cx43 and Sox4 expression. The bottom panel focusing on the myocardial expression zone of Sox4 at the border of the left ventricle and outflow tract. la Left atrium, ra right atrium, lv left ventricle, rv right ventricle, oft outflow tract, cus cushion mesenchyme, myo myocardium
A potential downstream target of the Tbx3–Sox4 interaction
For many transcription factors, including T-box proteins, target promoter specificity may be achieved through interaction with other proteins [17, 18, 36, 37]. In several recent studies, we and others have addressed the functional role of T-box proteins, particularly Tbx3, in the development of AV and outflow regions of the heart [11, 12, 38, 39]. Complimentary to recent microarray experiments to determine the downstream targets of Tbx3 [10] (unpublished data, MLB, VMC), we have carried out a Tbx3 ChIP-seq experiment to identify direct gene targets and provide a genome-wide map of Tbx3-binding sites (complete dataset will be published elsewhere). The quality of the data generated by this ChIP-seq approach could be validated by the marked presence in the sequence peaks of several published T-box-binding sites and gene enhancer elements (Supplementary Fig. 1). Spurred by our novel finding of expression of Sox4 in the myocardium, we were intrigued by recent reports describing ChIP-binding experiments of SOX2 and SOX4 [25, 26]. Close examination of these datasets revealed an evolutionarily conserved region in the first intron of the Gja1 gene, that is bound by both SOX2 and SOX4. Repression of Gja1 in the heart is known to involve Tbx2 and Tbx3, which may display redundant roles in this process. Furthermore, myocardial Gja1 expression is complimentary to myocardial expression patterns of Tbx2 and Tbx3 [12, 19, 21], as well as the myocardial expression of Sox4 (Fig. 4). As shown in Fig. 5a, our Chip-seq data shows that the same region of Gja1 in intron 1 as found in the SOX2 and SOX4 ChIP experiments is also bound by Tbx3, implicating that it may be a conserved genomic element important for the regulation of Gja1. A transcription factor-binding site prediction using high quality position weight matrices (Jaspar database) yielded as many as 11 potential Sox-binding sites and 4 potential T-box-binding sites (Fig. 5a).
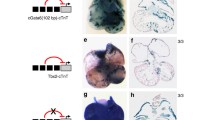
Regulation by T-box proteins and SOX4 of a putative Gja1 enhancer. Overlapping SOX2 ChIP-seq, SOX4 ChIP-chip, P300 ChIP-seq, and Tbx3 ChIP-seq data in intron 1 Gja1. a Visualized as UCSC custom tracks. Tbx3 data shows peak profiles for tags sequenced in hearts from Tbx3-induced mice. Predicted binding sites for T-box factors (open triangles) and Sox proteins (closed triangles) are indicated. b In vivo verification of Tbx3 and Sox4 association within this overlap (black line with arrow heads (Fig. 5a) marks the position of the target amplification, Gja1) using ChIP-QPCR. The result is presented as an enrichment relative to an IgG control. Amplification of the known T-box-binding site [36] within the proximal Nppa promoter is also shown. This region (supplementary Fig. 1) shows the expected enrichment for Tbx3, but no enrichment for Sox4, as based on the Tbx3 ChIP-seq and the SOX4 ChIP-chip data. c In vivo verification using ChIP-PCR of Tbx3, Sox4 and p300 association within this overlapping binding region. In this case, the negative controls are carried out using the same ChIP chromatin from the Tbx3, Sox4, and P300 IPs in combination with primers specific to a genomic region known not to bind Tbx3, Sox4, or P300. d In vivo analysis of zebrafish embryos (72 hpf) expressing H2AeGFP fusion protein under control of the minimal e1b promoter (Control) and minimal promoter + putative Gja1 enhancer (Enhancer). The genomic region used to generate this clone is marked with a solid green bar in Fig. 5a. Fish with this Gja1 enhancer construct clearly show restricted and specific expression of eGFP in the ventricle and atrium of the heart. V Ventricle, A atrium, P pericardium
A small element in intron 1 of Gja1 is occupied by Tbx3, Sox4, and P300 in vivo and drives expression in the vertebrate heart
Sox protein ChIP studies and our own mouse heart Tbx3 ChIP studies made use of different organisms and tissue types. Both Sox studies were carried out in human tissues, SOX2 making use of a ChIP-microarray approach in embryonic stem cells and SOX4, a ChIP-chip in a prostate cell line. We therefore first validated that both Tbx3 and Sox4 could occupy this element in the same system. To this end, a ChIP analysis was carried out using embryonic day 11.5 hearts isolated from wild-type mice. Using either anti-Tbx3 antibodies, anti-Sox4 antibodies, or matched IgG as control, both Tbx3 and Sox4 are found to occupy this region of Gja1, in vivo, at the same stage of mouse heart development (Fig. 5b). Since the region identified in Gja1 may represent an as yet unidentified enhancer element, we decided to test for P300 association. P300 is a ubiquitously expressed protein known to bind active enhancers across the genome [40]. Using ChIP–PCR (Fig. 5c), P300 can indeed be found to bind this region in vivo, an observation that is in agreement with P300 embryonic heart ChIP-seq data recently generated by Blow and coworkers [41] (Fig. 5a). To further validate that the Gja1 intronic element can function as an enhancer in vivo, we tested the expression of GFP under control of this element, using a zebrafish enhancer assay system. GFP expression in zebrafish can be found restricted to the heart and shows a confinement to cells of the ventricle and, albeit a lower level, the atrium (Fig. 5d). No expression was observed in control fish carrying the construct lacking the enhancer element. Fish expressing the enhancer-GFP construct also displayed pericardial edema, indicating a level of enhancer-construct toxicity.
Assessing the synergistic potential of the Tbx3–Sox4 interaction using the Gja1 enhancer element
To test the function of this enhancer element in-terms of the Tbx3–Sox4 complex, it was cloned upstream of a minimal E1b promoter sequence and tested for its ability to induce expression of a luciferase reporter gene in COS7 cells (Fig. 6). Significant up-regulation, 15-fold, of luciferase was observed using this construct when compared to the empty vector possessing the minimal promoter alone. Both Tbx2 and Tbx3 were able to significantly down-regulate expression of luciferase from this construct. Tbx5 has no significant effect when co-transfected. Addition of Sox4 alone resulted in an eightfold increase in luciferase expression. However, in the presence of Sox4, Tbx2, and Tbx3 displayed a significantly increased capacity to down-regulate this enhancer element. In this context, addition of Tbx5 had no significant effect on luciferase expression. These results indicate a competitive and yet synergistic transcription effect of Sox4 on both Tbx2 and Tbx3.
Transfection of luciferase under control of a minimal promoter and the Gja1 enhancer. Transfections of a Gja1 intronic enhancing region (marked with a solid green bar in Fig. 5a) reporter construct in the presence of Tbx2, Tbx3 or Tbx5 and SOX4. Addition of Tbx2 or Tbx3 alone results in an approximate 1.7-fold down-regulation of enhancing activity. SOX4 alone is able to up-regulate activity of this enhancer, but in the presence of Tbx2 and Tbx3 appears able to stimulate the down-regulation capacity of both Tbx2 and Tbx3. * and # denotes p < 0.0001
Discussion
Members of the T-box and Sox families of transcriptional regulators control a diverse array of processes during vertebrate embryonic development [13, 42]. In this study, we present evidence that Tbx3 and Sox4 interact via their DNA-binding domains, both in vitro and in mammalian cells. Comparative expression and ChIP analysis also demonstrates that this interaction may be functional at transcriptional regulation sites during development and that Sox4 may facilitate the transcriptional activities of Tbx3 at gene enhancer locations.
The interaction studies presented here show that the DNA-binding domains of Tbx3 and Sox4 interact. Since this interaction occurs through highly conserved domains, one might expect other members of the T-box family to be able to interact with Sox4. Indeed, the related proteins Tbx2 and Tbx5 also bind Sox4. The apparent lack of specificity within this closely related group of T-box factors is also evident for Nkx2.5 and Gata4, which partner-up with multiple T-box genes (reviewed in [43]). The functionality of these interactions is likely dictated by the timing and (co-) localization of expression and the relative expression levels of the different T-box factors.
In relation to the specific molecular function and significance of the T-box–Sox interaction we describe here, Sox proteins appear to predominantly function as transcriptional activators [44, 45], often serving to position gene enhancers, by DNA bending and opening [46], in a more fortuitous position for functional interaction of other transactivating factors. In this respect, addition of Sox4 in our transfection assays agrees with this statement, though at the same time the activities of Tbx3 and Tbx2, serving to down regulate transactivation, also appears to be facilitated by the presence of Sox4. It is interesting that Sox proteins are known to interact with a wide range of transcription factors [47] and as such may function here in facilitation of a transcriptional response based on the factor(s) present. Therefore the total transcriptional response of a gene or set of genes is not being driven by an individual protein, but by the stoichiometry and make-up of the complex of which it is a member.
We show that the C-terminal part of the HMG domain is essential for the interaction between Tbx3 and Sox4. Protein–protein interactions of Sox2, Sox8 and Sox10 with other transcription factors were also shown to be mediated by the C-terminal part of the HMG domain, which includes helix 3 and the C-terminal tail region [47–49]). These regions are not involved directly in establishing DNA contacts and are still available for interactions with other proteins even when Sox proteins are DNA bound [48, 50]. Similarly, the high degree of sequence conservation between HMG domains suggests that other members of the Sox family may also interact with Tbx3 [49]. For instance, the very early expression of Tbx3 in the inner cell mass of the blastocyst, where related T-box factors are not yet expressed, coincides with Sox2 expression, thus representing an example of a potentially interesting interaction worth further investigation [23, 51]. This statement seems particularly prudent in light of the recent publication suggesting a role for Tbx3 alongside Sox2 in maintaining stem cell pluripotency during embryonic stem cell development [52].
The novel finding of Sox4 in a small localized region of the interventricular ring myocardium, raises the question of a specific function for Sox4 in the myocardium in this region of the heart. Although Tbx3 and Tbx2 expression overlap with Sox4 in this region of the myocardium and may thus form a regulatory complex, by far the most extensive co-expression in the heart is seen in the mesenchyme of the atrioventricular region and outflow tract. This area is most likely the origin of the observed outflow tract malformations and early death observed in Sox4 knock out mice [28]. In this respect, and in terms of defining a possible functional role for a T-box–Sox interaction, the identification of Gja1 as direct downstream target for both Sox4 and Tbx3, is of particular relevance in terms of outflow tract development [12, 28, 53, 54] and the development of other tissues and organs such as the limbs [55, 56]. In humans, mutations in Gja1 give rise to the autosomal dominant disease oculodentodigital dysplasia (ODDD) (OMIM #164200) affecting the face, eyes, teeth, and limbs, and is associated with cardiac arrhythmia and neurological disorders. In mice, a model for ODDD has been generated by mutating Gja1. These mice display a phenotype overlap with the human disease, including syndactyly and cardiac arrhythmias. Altered expression of Cx43 has also been linked to numerous defects, including cardiac conduction [57] and mouse models have demonstrated that a critical regulation of Cx43 is necessary for correct outflow tract development, with over-expression and knockout models resulting in malformations [54, 58, 59].
Tbx3 (and Tbx2) knockout studies show clear ectopic expression of Cx43, revealing their key role in Gja1 regulation [10, 12, 38]. Interestingly, knockout studies of the closely related T-box factor, Tbx5, have suggested that Tbx5 plays no role in the regulation of Gja1 [36] and the data we present here would seem to support this statement. Previous studies relating to regulation of Gja1 by T-box proteins have focused predominantly on upstream regions shown to drive aspects of Cx43 expression [21, 60, 61]. However, to date, the elements that truly drive and control cardiac Gja1 expression, in vivo, have not been identified. Whereas the proximal promoter region does contain conserved transcription factor binding sites that can be functional in the repression or induction of reporter constructs in vitro, the 7-kb proximal promoter is not sufficient to drive expression in the heart [62]. Studies using this proximal promoter region show that it does mark the neural crest population which migrates to the outflow tract of the heart where it populates the cushions and appears to play some, as yet unknown, role in the septation of the outflow tract and patterning of the aortic arch region [63].
Here we present evidence of a regulatory enhancer positioned within intron 1 of Gja1, that we initially identify on the basis of a localized binding affinity for Tbx3, Sox2, Sox4, and P300. This multi-factorial binding coupled with an in vivo expression study in zebrafish provides strong evidence that this region is a functional enhancer during vertebrate heart development, which may be subject to strict spatiotemporal regulation by various T-box complexes including the T-box–Sox complex we describe here. Further, this novel enhancer element that we identify in intron 1 of Gja1 seems to contain sequences that induce expression of Gja1 in the developing heart. It is also interesting to note that the expression of GFP we observe in zebrafish, driven by this element, appears restricted to the atrium and ventricle and is apparently absent from the atrioventricular region. Recent studies tracing the expression of Tbx2 and Tbx3 isoforms in the zebrafish heart have shown a restriction of these factors to the atrioventricular region of the heart after approximately 33 hpf [64, 65]. This seems to suggest that the element we identify shows a level of functional as well as structural conservation. In line with this hypothesis, we have initiated a study devoted to investigating and dissecting the specific function of this enhancer in the developing mouse heart to further address these issues.
References
Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE (2005) T-Box genes in vertebrate development. Annu Rev Genet 39:219–239
Packham EA, Brook JD (2003) T-box genes in human disorders. Hum Mol Genet 12:R37–R44
Linden H, Williams R, King J, Blair E, Kini U (2009) Ulnar mammary syndrome and TBX3: expanding the phenotype. Am J Med Genet A 149A:2809–2812
Jacobs JJL, Keblusek P, Robanus Maandag E, Kristel P, Lingbeek M, Nederlof PM, van Welsem T, van de Vijver MJ, Koh EY, Daley GQ, van Lohuizen M (2000) Senescence bypass screen identifies Tbx2, which represses Cdkn2a (p19 ARF) and is amplified in a subset of human breast cancers. Nat Genet 26:291–299
Prince S, Carreira S, Vance KW, Abrahams A, Goding CR (2004) Tbx2 directly represses the expression of the p21(WAF1) cyclin-dependent kinase inhibitor. Cancer Res 64:1669–1674
Fan W, Huang X, Chen C, Gray J, Huang T (2004) TBX3 and its isoform TBX3+2a are functionally distinctive in inhibition of senescence and are overexpressed in a subset of breast cancer cell lines. Cancer Res 64:5132–5139
Lingbeek ME, Jacobs JJ, van Lohuizen M (2002) The T-box repressors TBX2 and TBX3 specifically regulate the tumor suppressor gene p14ARF via a variant T-site in the initiator. J Biol Chem 277:26120–26127
Brummelkamp TR, Kortlever RM, Lingbeek M, Trettel F, MacDonald ME, van Lohuizen M, Bernards R (2002) TBX-3, the gene mutated in Ulnar-mammary syndrome, is a negative regulator of p19ARF and inhibits senescence. J Biol Chem 277:6567–6572
Yarosh W, Barrientos T, Esmailpour T, Lin L, Carpenter PM, Osann K, nton-Culver H, Huang T (2008) TBX3 is overexpressed in breast cancer and represses p14 ARF by interacting with histone deacetylases. Cancer Res 68:693–699
Hoogaars WM, Engel A, Brons JF, Verkerk AO, de Lange FJ, Wong LY, Bakker ML, Clout DE, Wakker V, Barnett P, Ravesloot JH, Moorman AF, Verheijck EE, Christoffels VM (2007) Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev 21:1098–1112
Mesbah K, Harrelson Z, Theveniau-Ruissy M, Papaioannou VE, Kelly RG (2008) Tbx3 is required for outflow tract development. Circ Res 103:743–750
Bakker ML, Boukens BJ, Mommersteeg MTM, Brons JF, Wakker V, Moorman AFM, Christoffels VM (2008) Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circ Res 102:1340–1349
Hoogaars WMH, Barnett P, Moorman AFM, Christoffels VM (2007) T-box factors determine cardiac design. Cell Mol Life Sci 64:646–660
Christoffels VM, Habets PEMH, Franco D, Campione M, de Jong F, Lamers WH, Bao ZZ, Palmer S, Biben C, Harvey RP, Moorman AFM (2000) Chamber formation and morphogenesis in the developing mammalian heart. Dev Biol 223:266–278
Christoffels VM, Hoogaars WMH, Tessari A, Clout DEW, Moorman AFM, Campione M (2004) T-box transcription factor Tbx2 represses differentiation and formation of the cardiac chambers. Dev Dyn 229:763–770
Mommersteeg MTM, Hoogaars WMH, Prall OWJ, de Gier-de Vries C, Wiese C, Clout DEW, Papaioannou VE, Brown NA, Harvey RP, Moorman AFM, Christoffels VM (2007) Molecular pathway for the localized formation of the sinoatrial node. Circ Res 100:354–362
Habets PEMH, Moorman AFM, Clout DEW, van Roon MA, Lingbeek M, Lohuizen M, Campione M, Christoffels VM (2002) Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev 16:1234–1246
Hiroi Y, Kudoh S, Monzen K, Ikeda Y, Yazaki Y, Nagai R, Komuro I (2001) Tbx5 associates with Nkx2–5 and synergistically promotes cardiomyocyte differentiation. Nat Genet 28:276–280
Hoogaars WMH, Tessari A, Moorman AFM, de Boer PAJ, Hagoort J, Soufan AT, Campione M, Christoffels VM (2004) The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc Res 62:489–499
Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, Thiessen N, Griffith O, He A, Marra M, Snyder M, Jones S (2007) Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methonds 4:651–657
Boogerd KJ, Wong LYE, Christoffels VM, Klarenbeek M, Ruijter JM, Moorman AFM, Barnett P (2008) Msx1 and Msx2 are functional interacting partners of T-box factors in the regulation of connexin 43. Cardiovasc Res 78:485–493
Moorman AFM, Houweling AC, de Boer PAJ, Christoffels VM (2001) Sensitive nonradioactive detection of mRNA in tissue sections: novel application of the whole-mount in situ hybridization protocol. J Histochem Cytochem 49:1–8
Chapman DL, Garvey N, Hancock S, Alexiou M, Agulnik SI, Gibson-Brown JJ, Cebra-Thomas J, Bollag RJ, Silver LM, Papaioannou VE (1996) Expression of the T-box family genes, Tbx1–Tbx5, during early mouse development. Dev Dyn 206:379–390
Sohal DS, Nghiem M, Crackower MA, Witt SA, Kimball TR, Tymitz KM, Penninger JM, Molkentin JD (2001) Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res 89:20–25
Scharer CD, McCabe CD, li-Seyed M, Berger MF, Bulyk ML, Moreno CS (2009) Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells. Cancer Res 69:709–717
Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122:947–956
Liao Y-L, Sun Y-M, Chau G-Y, Chau Y-P, Lai T-C, Wang J-L, Horng J-T, Hsiao M, Tsou A-P (2008) Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene 27:5578–5589
Ya J, Schilham MW, de Boer PAJ, Moorman AFM, Clevers H, Lamers WH (1998) Sox4-deficiency syndrome in mice is an animal model for common trunk. Circ Res 83:986–994
Goldsworthy M, Hugill A, Freeman H, Horner E, Shimomura K, Bogani D, Pieles G, Mijat V, Arkell R, Bhattacharya S, Ashcroft FM, Cox RD (2008) Role of the transcription factor Sox4 in insulin secretion and impaired glucose tolerance. Diabetes 57:2234–2244
Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PAJ, Verbeek S, Lamers WH, Kruisbeek AM, Cumano A, Clevers H (1996) Sox-4 gene is required for cardiac outflow tract formation and pro-B lymphocyte expansion. Nature 380:711–714
Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, Weese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Tasneem A, Thanki N, Yamashita RA, Zhang D, Zhang N, Bryant SH (2009) CDD: specific functional annotation with the conserved domain database. Nucleic Acids Res 37:D205–D210
Hur EH, Hur W, Choi JY, Kim IK, Kim HY, Yoon SK, Rhim H (2004) Functional identification of the pro-apoptotic effector domain in human Sox4. Biochem Biophys Res Commun 325:59–67
Dy P, Penzo-Mendez A, Wang H, Pedraza CE, Macklin WB, Lefebvre V (2008) The three SoxC proteins–Sox4, Sox11 and Sox12–exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res 36:3101–3117
Hoogaars WMH, Barnett P, Rodriguez M, Clout DE, Moorman AFM, Goding CR, Christoffels VM (2008) TBX3 and its splice variant TBX3+exon 2a are functionally similar. Pigment Cell Melanoma Res 21:379–387
Wessels A, Vermeulen JLM, Verbeek FJ, Virágh Sz, Kálmán F, Lamers WH, Moorman AFM (1992) Spatial distribution of “tissue-specific” antigens in the developing human heart and skeletal muscle: III. An immunohistochemical analysis of the distribution of the neural tissue antigen G1N2 in the embryonic heart; implications for the development of the atrioventricular conduction system. Anat Rec 232:97–111
Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, Seidman JG (2001) A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 106:709–721
Black BL, Olson EN (1998) Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol 14:167–196
Aanhaanen WT, Brons JF, Dominguez JN, Rana MS, Norden J, Airik R, Wakker V, de Gier-de Vries C, Brown NA, Kispert A, Moorman AF, Christoffels VM (2009) The Tbx2+ primary myocardium of the atrioventricular canal forms the atrioventricular node and the base of the left ventricle. Circ Res 104:1267
Dupays L, Kotecha S, Mohun TJ (2009) Tbx2 misexpression impairs deployment of second heart field derived progenitor cells to the arterial pole of the embryonic heart. Dev Biol 333:121–131
Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA (2009) ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457:854–858
Blow M, McCulley D, Li Z, Zhang T, Akiyama J, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Bristow J, Ren B, Black B, Rubin E, Visel A, Pennacchio L (2010) ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet 42
Restivo A, Piancentini G, Placidi S, Saffirio C, Marino B (2006) Cardiac outflow tract: a review of some embryogenetic aspects of the conotruncal region of the heart. Anat Rec A Discov Mol Cell Evol Biol 288A:936–943
Boogerd CJ, Moorman AF, Barnett P (2009) Protein interactions at the heart of cardiac chamber formation. Ann Anat 191:505–517
Wegner M (2009) All purpose Sox: The many roles of Sox proteins in gene expression. Int J Biochem Cell Biol 42:381–390
Aaboe M, Birkenkamp-Demtroder K, Wiuf C, Sorensen FB, Thykjaer T, Sauter G, Jensen KM, Dyrskjot L, Orntoft T (2006) SOX4 expression in bladder carcinoma: clinical aspects and in vitro functional characterization. Cancer Res 66:3434–3442
Ferrari S, Harley VR, Pontiggia A, Goodfellow PN, Lovell-Badge R, Bianchi ME (1992) SRY, like HMG1, recognizes sharp angles in DNA. EMBO J 11:4497–4506
Wissmuller S, Kosian T, Wolf M, Finzsch M, Wegner M (2006) The high-mobility-group domain of Sox proteins interacts with DNA-binding domains of many transcription factors. Nucleic Acids Res 34:1735–1744
Remenyi A, Lins K, Nissen LJ, Reinbold R, Scholer HR, Wilmanns M (2003) Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev 17:2048–2059
Kamachi Y, Uchikawa M, Tanouchi A, Sekido R, Kondoh H (2001) Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev 15:1272–1286
Werner MH, Huth JR, Gronenborn AM, Clore GM (1995) Molecular basis of human 46X, Y sex reversal revealed from the three-dimensional solution structure of the human SRY–DNA complex. Cell 81:705–714
Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R (2003) Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 17:126–140
Han J, Yuan P, Yang H, So BS, Li P, Lim SL, Cao S, Tay J, Orlov Y, Lufkin T, Ng HH, Tam W, Lim B (2010) Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature doi:10.1038/nature08735
Liu S, Liu F, Schneider AC, St.Amand T, Epstein JA, Gutstein DE (2006) Distinct cardiac malformations caused by absence of connexin 43 in the neural crest and in the non-crest neural tube. Dev 133:2063–2073
Huang GY, Wessels A, Smith BR, Linask KK, Ewart JL, Lo CW (1998) Alteration in connexin 43 gap junction gene dosage impairs conotruncal heart development. Dev Biol 198:32–44
Kalcheva N, Qu J, Sandeep N, Garcia L, Zhang J, Wang Z, Lampe PD, Suadicani SO, Spray DC, Fishman GL (2007) Gap junction remodeling and cardiac arrhythmogenesis in a murine model of oculodentodigital dysplasia. Proc Natl Acad Sci USA 104:20512–20516
Dobrowolski R, Hertig G, Lechner H, Wörsdörfer P, Wulf V, Dicke N, Eckert D, Bauer R, Schorle H, Willecke K (2009) Loss of connexin43-mediated gap junctional coupling in the mesenchyme of limb buds leads to altered expression of morphogens in mice. Hum Mol Genet 18:2899–2911
Danik SB, Liu F, Zhang J, Suk HJ, Morley GE, Fishman GI, Gutstein DE (2004) Modulation of cardiac gap junction expression and arrhythmic susceptibility. Circ Res 95:1035–1041
Li WE, Waldo K, Linask KL, Chen T, Wessels A, Parmacek MS, Kirby ML, Lo CW (2002) An essential role for connexin43 gap junctions in mouse coronary artery development. Dev 129:2031–2042
Xu X, Francis R, Wei CJ, Linask KL, Lo CW (2006) Connexin 43-mediated modulation of polarized cell movement and the directional migration of cardiac neural crest cells. Dev 133:3629–3639
Chen JR, Chatterjee B, Meyer R, Yu JC, Borke JL, Isales CM, Kirby ML, Lo CW, Bollag RJ (2004) Tbx2 represses expression of connexin43 in osteoblastic-like cells. Calcif Tissue Int 74:561–573
Lo CW, Cohen MF, Huang GY, Lazatin BO, Patel N, Sullivan R, Pauken C, Park SM (1997) Cx43 gap junction gene expression and gap junctional communication in mouse neural crest cells. Dev Genet 20:119–132
Chatterjee B, Chin A, Valdimarsson G, Finis C, Sonntag J, Choi B, Tao L, Balasubramanian K, Bell C, Krufka A, Kozlowski D, Johnson R, Lo C (2005) Developmental regulation and expression of the zebrafish connexin43 gene. Dev Dyn 233:890–906
Waldo K, Zdanowicz M, Burch J, Kumiski DH, Stadt HA, Godt RE, Creazzo TL, Kirby ML (1999) A novel role for cardiac neural crest in heart development. J Clin Invest 103:1499–1507
Camarata T, Krcmery J, Snyder D, Park S, Simon H (2010) Pdlim7 (LMP4) regulation of Tbx5 specifies zebrafish heart atrio-ventricular boundary and valve formation. Dev Biol 337:233–245
Ribeiro I, Kawakami Y, Buscher D, Raya A, Rodriguez-Leon J, Morita M, Rodriguez Esteban C, Izpisua Belmonte JC (2007) Tbx2 and Tbx3 regulate the dynamics of cell proliferation during heart remodeling. PLoS ONE 2:e398
Acknowledgments
This project was funded by the Netherlands Heart Foundation Grant 1996M002 and European Community’s Sixth Framework Programme contract HeartRepair LSHM-CT-2005-018630. We would like to thank J.M. Ruijter for his assistance with statistical analysis.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
C. J. J. Boogerd and L. Y. E. Wong have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Boogerd, C.J.J., Wong, L.Y.E., van den Boogaard, M. et al. Sox4 mediates Tbx3 transcriptional regulation of the gap junction protein Cx43. Cell. Mol. Life Sci. 68, 3949–3961 (2011). https://doi.org/10.1007/s00018-011-0693-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-011-0693-7