Abstract
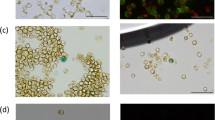
A nuclear transformation system has been developed for the diatomPhaeodactylum tricornutum using microparticle bombardment to introduce thesh ble gene fromStreptoalloteichus hindustanus into cells. Thesh ble gene encodes a protein that confers resistance to the antibiotics Zeocin and phleomycin. Chimeric genes containing promoter and terminator sequences from theP. tricornutum fcp genes were used to drive expression ofsh ble. Between 10–100 transformants were recovered/108 cells. Transformants were able to grow on at least 500 µg/ml of Zeocin, which is 10 times the amount necessary to kill wild-type cells. Based on Southern hybridizations thesh ble gene was present in 1–3 copies/transformant. Relative levels of correctly processed transcripts were correlated with the abundance of the Sh ble protein (present at 0.1–2.0 µg/mg total protein). Thecat reporter gene fused to afcp promoter could also be introduced by microparticle bombardment and was found to be highly expressed (average of 7.1 U/mg total protein). This work demonstrates that heterologous genes can be readily expressed inP. tricornutum. The development of selectable marker and reporter gene constructs provides the tools necessary for dissecting gene structure and regulation, and introducing novel functions into diatoms.
Similar content being viewed by others
References
Alwyn T, Rees V (1995) On ammonia futile cycling in a marine unicellular alga. Biochem Biophys Acta 1228:254–260
Apt KE, Bhaya D, Grossman AR (1994) Characterization of genes encoding the light-harvesting proteins in diatoms: biogenesis of the fucoxanthin chlorophylla/c protein complex. J Appl Phycol 6:225–230
Apt KE, Sukenik A, Grossman R (1995) The ER chaperone BiP from the diatomPhaeodactylum tricornutum (PGR95-061). Plant Physiol 109:339
Armaleo D, Ye G-N, Klein TM, Shark KB, Sanford JC, Johnston SA (1990) Biolistic nuclear transformation ofSaccharomyces cerevisiae and other fungi. Curr Genet 17:97–103
Barclay WR, Meager KM, Abril JR (1994) Heterotrophic production of long chain omega-3 fatty acids utilizing algae and algae-like organisms. J Appl Phycol 6:123–129
Berdy J (1980) Bleomycin-type Antibiotics. In: Berdy J (ed) Handbook of antibiotic compounds, vol. IV, Part I. Amino acid and peptide antibiotics. CRC Press, Boca Raton, pp 459–497
Bhaya D, Grossman AR (1993) Characterization of gene clusters encoding the fucoxanthin chlorophyll proteins of the diatomPhaeodactylum tricornutum. Nucleic Acids Res 21:4458–4466
Chomczynski P (1994) Single-step method of total RNA isolation by acid guanidine-phenol extraction. In: Celis JE (ed) Cell biology: a laboratory handbook, vol I. Academic Press, San Diego, pp 680–683
de la Luna S, Ortin J (1992)pac gene as an efficient dominant marker and reporter gene in mammalian cells. Methods Enzymol 216:376–385
Drocourt D, Calmels TPG, Reynes JP, Baron M, Tiraby G (1990) Cassettes of theStreptoalloteichus hindustanus ble gene for transformation of lower and higher eukaryotes to phleomycin resistance. Nucleic Acids Res 18:4009
Dunahay TG, Jarvis EE, Roessler PG (1995) Genetic transformation of the diatomsCyclotella cryptica andNavicula saprophila. J Phycol 31:1004–1012
Galloway RE (1990) Selective conditions and isolation of mutants in salt-tolerant lipid-producing microalgae. J Phycol 26:752–760
Gerwick WH, Roberts MA, Proteau PJ, Chen J-L (1994) Screening cultured marine microalgae for anticancer-type activity. J Appl Phycol 6:143–150
Gladue RM, Maxey JE (1994) Microalgal feeds for aguaculture. J Appl Phycol 6:131–141
Goff LJ, Coleman AW (1988) The use of plastid DNA restriction endonuclease patterns in delineating red algal species and populations. J Phycol 24:357–368
Gold SE, Bakkeren G, Davis JE, Kronstad JW (1994) Three selectable markers for transformation ofUstilago maydis. Gene 142:225–230
Guillard RRL (1975) Culture of phytoplankton for feeding marine invertebrates. In: Smith WL, Chanley MH (eds) Culture of marine invertebrate animals. Plenum Press, New York, pp 26–60
Haughn GW, Somerville C (1986) Sulfonylurea-resistant mutants ofArabidopsis thaliana. Mol Gen Genet 204:430–434
Ianora A, Poulet SA, Miralto A (1995) A comparative study on the inhibitory effect of diatoms on the reproductive biology of the copepodTemora stylifera. Mar Biol 121:533–539
Khalyfa A, Kermasha S, Marsot P, Goetghebeur M (1995) Purification and characterization of chlorophyllase from the algaPhaeodactylum tricornutum by preparative native electrophoresis. Appl Biochem Biotech 53:11–27
Kindle KL, Skchell RA, Fernandez E, Lefebvre PA (1989) Stable nuclear transformation ofChlamydomonas using theChlamydomonas gene for nitrate reductase. J Cell Biol 109:2589–2601
Kuma K, Matsunga K (1995) Availability of colloidal ferric oxides to coastal marine phytoplankton. Marine Biol 122:1–11
Laemmli UK (1970) Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227:680–685
La Roche J, Murray H, Orellana M, Newton J (1995) Flavodoxin expression as an indirect indicator of iron limitation in marine diatoms. J Phycol 31:520–530
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Overbeek PA, Lai S-P, Van Quill KR, Westphal H (1986) Tissue-specific expression in transgenic mice of a fused gene containing RSV terminal sequences. Science 231:1574–1577
Rees TAV, Larson TR, Heldeus JWG, Huning FGJ (1995) In situ glutamine synthetase activity in a marine unicellular alga. Development of a sensitive colorimetric assay and the effects of nitrogen status on enzyme activity. Plant Physiol 109:1405–1410
Rosen KM, Lamperti ED, Villa-Komaroff L (1990) Optimizing the Northern blot procedure. Biotechniques 8:398–403
Round FE, Crawford RM, Mann DG (1990) The Diatoms: biology and morphology of the genera. Cambridge Univ Press, Cambridge, UK
Starr RC, Zeikus JA (1993) UTEX: the culture collection of algae at the University of Texas at Austin. J Phycol 29(Suppl):93
Ward A, Etessami P, Stanley J (1988) Expression of a bacterial gene in plants mediated by infectious geminivirus DNA. EMBO J 7:1583–1587
Werner D (1977) The biology of diatoms. Univ of California Press, Berkeley
Zhukova NN, Aizdaischer NA (1995) Fatty acid composition of 15 species of marine microalgae. Phytochemistry 39:351–356
Author information
Authors and Affiliations
Additional information
Communicated by R. G. Herrmann
Rights and permissions
About this article
Cite this article
Apt, K.E., Grossman, A.R. & Kroth-Pancic, P.G. Stable nuclear transformation of the diatomPhaeodactylum tricornutum . Molec. Gen. Genet. 252, 572–579 (1996). https://doi.org/10.1007/BF02172403
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02172403