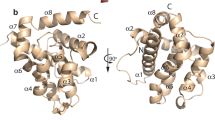
Abstract
The nature of the bcl-2 family of protooncogenes was analyzed by sequence alignment, secondary structure prediction, and phylogenetic techniques. Phylogenies were inferred from both the nucleic acid and amino acid sequences of the human, murine, rat, and chicken sequences for BCL-2 and BCL-X, human MCL1, murine A1, the nematode Caenorhabditis elegans and Caenorhabditis briggsiae ced-9 proteins, and the sequences BHRF1 from Epstein-Barr and LMW5-HL from African swine fever viruses. Both sequence alignment and secondary structure prediction techniques supported the conservation of both the overall secondary structure and the carboxy-terminal transmembrane domain in all members of the family. All the treeing methods employed (distance matrix, maximum likelihood, and parsimony) supported a tree in which the proapoptotic proteins BCL-2 and BCL-X represent the most recent additions to the group. All the trees also indicated that the viral proteins BHRF1 and LMW-HL arose from a common ancestor, an ancestor they shared in common with the pro-apoptotic control protein BAX, indicating that this function of BAX evolved only recently. The most ancient branches are represented by the nematode ced-9 protein and by the control genes MCL1 and A1, which in the treeing methods employed represent separate lineages within the most ancient grouping. These results demonstrate the evolution of a highly conserved family of developmental control genes from nematode to man—genes that encode proteins essential for normal development but which are highly conserved in terms of predicted structure and possible cellular localization. The evolutionary analysis also indicates that the family may be even larger than originally predicted and that other members are waiting to be discovered.
Similar content being viewed by others
References
Baer R (1994) Bcl-2 breathes life into embryogenesis (1994) Am J Pathol 145:7–10
Boise LH, González-Garcia M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nuñez G, Thompson CB (1993) bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell 74:597–608
Borner C, Martinou I, Mattmann C, Irmler M, Schaerer E, Martinou J-C, Tschopp J (1994) The protein bcl-2α does not require membrane attachment, but two conserved domains to suppress apoptosis. J Cell Biol 126:1059–1068
Dayhoff MO (1979) Atlas of protein sequence and structure, Vol 5, Suppl 3, 1978. National Biomedical Research Foundation, Washington, DC
Diaconis P, Efron B (1983) Computer-intensive methods in statistics. Sci Am 248:116–130
Felsenstein J (1993) Phylogenetic inference program (PHYLIP) manual, version 3.5. University of Washington, Seattle.
Felsenstein J (1985) Confidence limits in phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM, Margoliash E (1967) Construction of phylogenetic trees. Science 15:279–284
Haldar S, Beatty C, Croce CM (1990) Bcl-2 alpha encodes a novel small molecular weight GTP binding protein. Advances in Enzyme Regulation 30:145–153
Henderson SA, Huen D, Rowe M, Dawson C, Johnson G, Rickinson A (1993) Epstein-Barr virus-coded BHRFI protein, a viral homologue of Bcl-2, protects human B cells from programmed cell death. Proc Natl Acad Sci USA 90:8479–8483
Hengartner MO, Horovitz HR (1994) C elegans cell survival gene ced-9 encodes a functional homolog of the mammalian protooncogene bcl-2. Cell 76:665–676
Higgins DA, Sharp PM (1988) Clustal: a package for performing multiple sequence alignments on a microcomputer. Gene 73:237–244
Hoffman B, Lieberman DA (1994) Molecular controls of apoptosis: differentiation/growth arrest primary response genes, protooncogenes and tumor suppressor genes as positive & negative modulators. Oncogene 9:1807–1812
Jin L, Nei M (1990) Limitations of the evolutionary parsimony method of phylogenetic analysis. Mol Biol Evol 7:82–102
Kane DJ, Sarafian TA, Anton R, Hahn H, Butler Gralla E, Selverstone Valentine J, Örd T, Bredesen DE (1993) Bcl-2 inhibition of neural death: decreased generation of reactive oxygen species. Science 262:1274–1277
Kimura M (1980) A simple method for estimating evolutionary rates of base substitution through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Koonin EV, Senkevich TG (1992) Evolution of thymidine and thymidylate kinases: the possibility of independent capture of TK genes by different groups of viruses. Virus Genes 6:187–196
Kozpaz KM, Yang T, Buchan HL, Zhou P, Craig RW (1993) MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc Natl Acad Sci USA 90:3516–3520
Lin EY, Orlofsky A, Berger MS, Prystowsky MB (1993) Characterization of A1, a novel hemopoietic-specific early-response gene with sequence similarity to bcl-2. J Immunol 151:1979–1988
Neilan JG, Lu Z, Afonso CL, Kutish GF, Sussman MD, Rock DL (1993) An African swine fever virus gene with similarity to the proto-oncogene bcl-2 and the Epstein-Barr virus gene BHRF1. J Virol 67:4391–4394
Nguyen M, Millar DG, Young VW, Korsmeyer SJ, Shore GC (1993) Targeting of Bcl-2 to the mitochondrial outer membrane by a COOH-terminal signal anchor sequence. J Biol Chem 268:25265–25268
Oltvai ZN, Milliman CL, Korsmeyer SJ (1993) Bcl-2 heterodimerizes in vivo with a conserved homolog BAX that accelerates programmed cell death. Cell 74:609–619
Pai FF, Kabsch W, Krengel U, Holmes KG, John J, Wittinghofer A (1989) Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature 341:209–214
Reed JC (1994) Bcl-2 and the regulation of programmed cell death. J Cell Biol 124:1–6
Rost B, Sander C (1994) Combining evolutionary information and neural networks to predict protein secondary structure. Proteins 19:55–72 (press).
Saitou N, Nei M (1987) The neighbor joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Savaste M, Sibbald PR, Wittinghofer A (1990) The P-loop—a common motif in ATP-and GTP-binding proteins. Trends Biochem Sci 15: 430–434
Sipos L, von Heijne G (1993) Predicting the topology of eukaryotic membrane protein. Eur J Biochem 213:1333–1340
Veis Novack D, Kursmeyer SJ (1994) Bcl-2 protein expression during murine development. Am J Pathol 145:61–73
Zuckerkandl E (1987) On the molecular evolutionary clock. J Mol Evol 26:36–46
Author information
Authors and Affiliations
Additional information
Correspondence to: D. Lloyd Evans
Rights and permissions
About this article
Cite this article
Evans, D.L., Mansel, R.E. Molecular evolution and secondary structural conservation in the B-cell lymphoma leukemia 2 (bcl-2) family of proto-oncogene products. J Mol Evol 41, 775–783 (1995). https://doi.org/10.1007/BF00173157
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00173157