Fungal Immunomodulatory Protein from Nectria haematococca Suppresses Growth of Human Lung Adenocarcinoma by Inhibiting the PI3K/Akt Pathway
Abstract
:1. Introduction
2. Results
2.1. FIP-nha Inhibits Lung Adenocarcinoma Cell Growth Ex Vivo and In Vivo
2.2. Proteomic Analysis of FIP-Nha-Induced Differential Expressed Proteins in A549 Cells
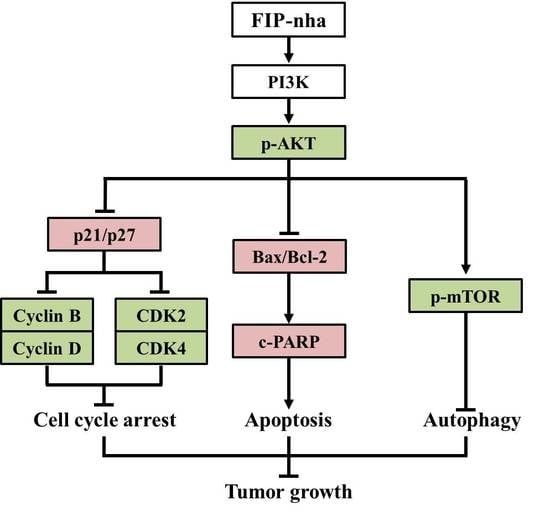
2.3. FIP-Nha Negatively Regulates the PI3K/Akt Signaling Pathway and Induces Cell Autophagy
2.4. FIP-Nha Induces G1/S and G2/M Cell Cycle Arrest in Lung Adenocarcinoma Cells
2.5. FIP-Nha Promotes Apoptosis in Lung Adenocarcinoma Cells
2.6. FIP-Nha Affects Cell Cycle, Autophagy, and Apoptosis Processes in Xenograft Mouse Tumors
3. Discussion
4. Materials and Methods
4.1. Materials and Chemicals
4.2. FIP-Nha Protein Expression and Purification
4.3. Cell Viability Measurement
4.4. In Vivo Tumor Xenograft Mouse Model
4.5. iTRAQ Proteomic Analysis
4.6. Western Blotting
4.7. TEM
4.8. Apoptosis Assays
4.9. DNA Content/Cell Cycle Analysis
4.10. IHC Analysis
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| FIP | Fungal immunomodulatory protein |
| FIP-nha | FIP from Nectria haematococca |
| FIP-fve | FIP from Flammulina velutipes |
| FIP-gmi | FIP from Ganoderma Microsporum |
| FIP-gts | FIP from Ganoderma tsugae |
| H2347 | NCI-H2347 (H2347) |
| IHC | Immunohistochemistry |
| iTRAQ | Isobaric tags for relative and absolute quantification |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LC3B | Microtubule-associated protein light chain B |
| LZ-8 | FIP from Ling Zhi-8 |
| mTOR | Mammalian target of rapamycin |
| NSCLC | Non-small cell lung cancer |
| p-Akt | Phosphorylated Akt |
| PI | Propidium iodide |
| RP | Reversed phase chromatography |
| SDS | Sodium dodecyl sulfate |
| TEM | Transmission electron microscope |
References
- Torre, L.A.; Siegel, R.L.; Jemal, A. Lung Cancer and Personalized Medicine; Springer: Heidelberg, Germany, 2016; p. 893. [Google Scholar] [CrossRef]
- Godoy, C.B.; Naidich, D.P. Subsolid pulmonary nodules and the spectrum of peripheral sdenocarcinomas of the lung: Recommended interim guidelines for assessment and management. Radiology. 2009, 253, 606–622. [Google Scholar] [CrossRef] [PubMed]
- Woodard, G.A.; Jones, K.D.; Jablons, D.M. Lung cancer staging and prognosis. Cancer Treat. Res. 2016, 170, 47–75. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Mao, Y.; You, Q.; Hua, D.; Cai, D. Piperlongumine induces apoptosis and autophagy in human lung cancer cells through inhibition of PI3K/Akt/mTOR pathway. Int. J. Immunopathol. Pharmacol. 2015, 28, 362–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prakash, O.; Kumar, A.; Kumar, P.; Ajeet, A. Anticancer Potential of Plants and Natural Products: A Review. Am. J. Pharmacol. Sci. 2013, 1, 104–115. [Google Scholar] [CrossRef]
- Shanmugam, M.K.; Warrier, S.; Kumar, A.P.; Sethi, G.; Arfuso, F. Potential role of natural compounds as anti-angiogenic agents in cancer. Curr. Vasc. Pharmacol. 2017, 15, 503–519. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981–2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Jie, S.; Hanchuan, D.; Moucheng, W. Characterization and immunomodulating activities of polysaccharide from Lentinus edodes. Int. Immunopharmacol. 2005, 5, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Xu, G.M.; Li, X.M.; Li, C.S.; Wang, B.G. Characterization of a Newly Isolated Marine Fungus Aspergillus dimorphicus for Optimized Production of the Anti-Tumor Agent Wentilactones. Mar. Drugs 2015, 13, 7040–7054. [Google Scholar] [CrossRef] [PubMed]
- Lai, I.C.; Lai, G.M.; Chow, J.M.; Lee, H.L.; Yeh, C.F.; Li, C.H.; Yan, J.L.; Chuang, S.E.; Whang-Peng, J.; Bai, K.J.; et al. Active fraction (HS7) from Taiwanofungus camphoratus inhibits AKT-mTOR, ERK and STAT3 pathways and induces CDK inhibitors in CL1-0 human lung cancer cells. Chin. Med. (UK) 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Prabavathy, D.; Nachiyar, C.V. Cytotoxicity of ethyl acetate extract of endophytic aspergillus fumigatus on A549 cell lines. Biosci. Biotechnol. Res. Asia 2014, 11, 797–802. [Google Scholar] [CrossRef]
- Kino, K.; Yamashita, A.; Yamaoka, K.; Watanabe, J.; Tanaka, S.; Ko, K.; Shimizu, K.; Tsunoo, H. Isolation and characterization of a new immunomodulatory protein, Ling Zhi-8 (LZ-8), from Ganoderma lucidium. J. Biol. Chem. 1989, 264, 472–478. [Google Scholar] [PubMed]
- Lin, W.H.; Hung, C.H.; Hsu, C.I.; Lin, J.Y. Dimerization of the N-terminal amphipathic α-helix domain of the fungal immunomodulatory protein from Ganoderma tsugae (Fip-gts) defined by a yeast two-hybrid system and site-directed mutagenesis. J. Biol. Chem. 1997, 272, 20044–20048. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.L.; Hsu, C.I.; Lin, R.H.; Kao, C.L.; Lin, J.Y. A New Fungal Immunomodulatory Protein, FIP-fve Isolated from the Edible Mushroom, Flammulina velutipes and its Complete Amino Acid Sequence. Eur. J. Biochem. 1995, 228, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Nie, Y.; Ding, Y.; Shi, L.; Tang, X. Recombinant expression of a novel fungal immunomodulatory protein with human tumor cell antiproliferative activity from Nectria haematococca. Int. J. Mol. Sci. 2014, 15, 17751–17764. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Jiang, Z.; Xu, W.; Xie, Y.; Zhao, L.; Tang, X.; Wang, F.; Xin, F. FIP-sch2, a new fungal immunomodulatory protein from Stachybotrys chlorohalonata, suppresses proliferation and migration in lung cancer cells. Appl. Microbiol. Biotechnol. 2017, 101, 3227–3235. [Google Scholar] [CrossRef] [PubMed]
- Hsin, I.L.; Ou, C.C.; Wu, T.C.; Jan, M.S.; Wu, M.F.; Chiu, L.Y.; Lue, K.H.; Ko, J.L. GMI, an immunomodulatory protein from Ganoderma microsporum, induces autophagy in non-small cell lung cancer cells. Autophagy 2011, 7, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.H.; Hsiao, Y.M.; Lin, C.H.; Yeh, C.S.; Wang, J.C.H.; Ni, C.H.; Hsu, C.P.; Ko, J.L. Induction of premature senescence in human lung cancer by fungal immunomodulatory protein from Ganoderma tsugae. Food Chem. Toxicol. 2008, 46, 1851–1859. [Google Scholar] [CrossRef] [PubMed]
- Cong, W.R.; Xu, H.; Liu, Y.; Li, Q.Z.; Li, W.; Zhou, X.W. Production and functional characterization of a novel fungal immunomodulatory protein FIP-SN15 shuffled from two genes of Ganoderma species. Appl. Microbiol. Biotechnol. 2014, 98, 5967–5975. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.W.; Hao, L.X.; Xu, G.X.; Sun, F.; Gao, F.; Zhang, R.; Liu, L.X. Molecular cloning and recombinant expression of a gene encoding a fungal immunomodulatory protein from Ganoderma lucidum in Pichia pastoris. World J. Microbiol. Biotechnol. 2009, 25, 383–390. [Google Scholar] [CrossRef]
- Patel, S.; Goyal, A. Recent developments in mushrooms as anti-cancer therapeutics: A review. 3 Biotech 2012, 2, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Martini, M.; De Santis, M.C.; Braccini, L.; Gulluni, F.; Hirsch, E. PI3K/AKT signaling pathway and cancer: An updated review. Ann. Med. 2014, 46, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Yip, P.Y. Phosphatidylinositol 3-kinase-AKT-mammalian target of rapamycin (PI3K-Akt-mTOR) signaling pathway in non-small cell lung cancer. Transl. Lung Cancer Res. 2015, 4, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Osaki, M.; Oshimura, M.; Ito, H. PI3K-Akt pathway: Its functions and alterations in human cancer. Apoptosis 2004, 9, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tang, N.; Hadden, T.J.; Rishi, A.K. Akt, FoxO and regulation of apoptosis. Biochim. Biophys. Acta Mol. Cell Res. 2011, 1813, 1978–1986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, N.; Dai, S.D.; Liu, N.N.; Liu, L.M.; Zhao, N.; Chen, L. PI3K/AKT/mTOR signaling pathway inhibitors in proliferation of retinal pigment epithelial cells. Int. J. Ophthalmol. 2012, 5, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Alonso, M.J.; Acosta, J.C.; Richard, C.; Delgado, M.D.; Sedivy, J.; León, J. p21Cip1 and p27Kip1 induce distinct cell cycle effects and differentiation programs in myeloid leukemia cells. J. Biol. Chem. 2005, 280, 18120–18129. [Google Scholar] [CrossRef] [PubMed]
- Downward, J. PI 3-kinase, Akt and cell survival. Semin. Cell Dev. Biol. 2004, 15, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yao, B.; Xu, M.; Zheng, X. RIP1 upregulation promoted tumor progression by activating AKT/Bcl-2/BAX signaling and predicted poor postsurgical prognosis in HCC. Tumor Biol. 2016, 37, 15305–15313. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.N.; Kumar, D.; Shankar, S.; Srivastava, R.K. Rottlerin induces autophagy which leads to apoptotic cell death through inhibition of PI3K/Akt/mTOR pathway in human pancreatic cancer stem cells. Biochem. Pharmacol. 2012, 84, 1154–1163. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.F.; Zhou, Z.W.; Li, X.X.; He, Z.; Pan, S.; Yang, Y.X.; Zhang, X.; Chew, K.; Yang, T.; Qiu, J.; et al. Induction of apoptosis and autophagy via sirtuin1- and PI3K/Akt/mTOR-mediated pathways by plumbagin in human prostate cancer cells. Drug Des. Dev. Ther. 2015, 9, 1511–1554. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.H.; Hsiao, Y.M.; Hsu, C.P.; Lin, M.Y.; Wang, J.C.H.; Huang, Y.L.; Ko, J.L. Transcriptionally mediated inhibition of telomerase of fungal immunomodulatory protein from Ganoderma tsugae in A549 human lung adenocarcinoma cell line. Mol. Carcinog. 2006, 45, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Mei, Y.; Sinha, S. Role of the crosstalk between autophagy and apoptosis in cancer. J. Oncol. 2013, 2013, 102735. [Google Scholar] [CrossRef] [PubMed]
- Hengartner, M.O. The biochemistry of apoptosis. Nature 2000, 407, 770–776. [Google Scholar] [CrossRef] [PubMed]
- Gump, J.M.; Thorburn, A. Autophagy and apoptosis: What is the connection? Trends Cell Biol. 2011, 21, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Degenhardt, K.; Mathew, R.; Beaudoin, B.; Bray, K.; Anderson, D.; Chen, G.; Mukherjee, C.; Shi, Y.; Gélinas, C.; Fan, Y.; et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 2006, 10, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Candia, M. Kenific and Jayanta Debnath Cellular and metabolic functions for autophagy in cancer cells. Trends Cell Biol. 2015, 25, 37–45. [Google Scholar] [CrossRef]
- Chen, Z.H.; Lam, H.C.; Jin, Y.; Kim, H.P.; Cao, J.; Lee, S.J.; Ifedigbo, E.; Parameswaran, H.; Ryter, S.W.; Choi, A.M.K. Autophagy protein microtubule-associated protein 1 light chain-3B (LC3B) activates extrinsic apoptosis during cigarette smoke-induced emphysema. Proc. Natl. Acad. Sci. USA 2010, 107, 18880–18885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, A.; Hu, R.; Zhao, Q.; Li, J.; Li, Y.; Yao, K.; Zhang, R.; Wang, H.; Yang, W.; Liu, Z. Autophagy induced by FTY720 promotes apoptosis in U266 cells. Eur. J. Pharm. Sci. 2012, 45, 600–605. [Google Scholar] [CrossRef] [PubMed]
- Shaid, S.; Brandts, C.H.; Serve, H.; Dikic, I. Ubiquitination and selective autophagy. Cell Death Differ. 2013, 20, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.C.; Hsiao, Y.M.; Wu, M.F.; Ou, C.C.; Lin, Y.W.; Lue, K.H.; Ko, J.L. Interruption of lung cancer cell migration and proliferation by fungal immunomodulatory protein FIP-fve from flammulina velutipes. J. Agric. Food Chem. 2013, 61, 12044–12052. [Google Scholar] [CrossRef] [PubMed]
- Ou, C.C.; Hsiao, Y.M.; Wang, W.H.; Ko, J.L.; Lin, M.Y. Stability of fungal immunomodulatory protein, FIP-gts and FIP-fve, in IFN-g production. Food Agric. Immunol. 2009, 20, 319–332. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M.; Wi, J.R. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Chen, L.; Lv, X.; Hou, G.; Wang, Y.; Jiang, C.; Zhu, H.; Xu, N.; Wu, L.; Lou, X.; et al. The levels of serine proteases in colon tissue interstitial fluid and serum serve as an indicator of colorectal cancer progression. Oncotarget 2016, 7, 32592–32606. [Google Scholar] [CrossRef] [PubMed] [Green Version]






© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, Y.; Li, S.; Sun, L.; Liu, S.; Wang, F.; Wen, B.; Sun, L.; Fang, X.; Chai, Y.; Cao, H.; et al. Fungal Immunomodulatory Protein from Nectria haematococca Suppresses Growth of Human Lung Adenocarcinoma by Inhibiting the PI3K/Akt Pathway. Int. J. Mol. Sci. 2018, 19, 3429. https://doi.org/10.3390/ijms19113429
Xie Y, Li S, Sun L, Liu S, Wang F, Wen B, Sun L, Fang X, Chai Y, Cao H, et al. Fungal Immunomodulatory Protein from Nectria haematococca Suppresses Growth of Human Lung Adenocarcinoma by Inhibiting the PI3K/Akt Pathway. International Journal of Molecular Sciences. 2018; 19(11):3429. https://doi.org/10.3390/ijms19113429
Chicago/Turabian StyleXie, Yingying, Shuying Li, Lei Sun, Shujun Liu, Fengzhong Wang, Boting Wen, Lichao Sun, Xiangdong Fang, Yushuang Chai, Hao Cao, and et al. 2018. "Fungal Immunomodulatory Protein from Nectria haematococca Suppresses Growth of Human Lung Adenocarcinoma by Inhibiting the PI3K/Akt Pathway" International Journal of Molecular Sciences 19, no. 11: 3429. https://doi.org/10.3390/ijms19113429