What Macromolecular Crowding Can Do to a Protein
Abstract
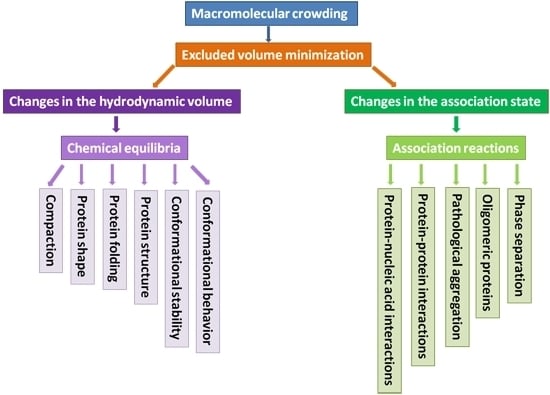
:1. Introduction

2. What Can Macromolecular Crowding Do to a Protein
2.1. How to Model Macromolecular Crowding?
| Macromolecular Crowders | Molecular Mass (kDa) | Hydrodynamic Radius (Å) | Effective Concentration a | Fractional vol. Occupancy Ψ b |
|---|---|---|---|---|
| Poly(ethylene glycol) PEG 2050 | 2 | 3.8 c/11.3 d | ||
| Dextran sulfate 10 e | 10 | <10 | ||
| Bovine pancreatic trypsin inhibitor (BPTI) f | 6.5 | 14.2 | ||
| PEG 4600 d | 4.6 | 17.9 | ||
| Ribonuclease A g | 13.7 | 19.3 | ||
| Lysozyme g | 14.3 | 20.0 | ||
| PEG 6000 d | 6.0 | 20.8 | ||
| PEG 8000 d | 8.0 | 24.5 | ||
| β-Lactoglobulin g | 36.8 | 27.1 | ||
| Hemoglobin g | 64.5 | 33.2 | ||
| Bovine serum albumin (BSA)e | 66.3 | 33.9 | 80 mg/mL | 18% |
| PEG 20000 | 20 | 34.5 h/41.4 d | ||
| Ficoll 70 e | 70 | 40 | 37.5 mg/mL | 17% |
| PEG 35000 d | 35 | 57.0 | ||
| Ficoll 400 e | 400 | 80 | 25 mg/mL | |
| Dextran 670 e | 670 | 210 | ||
| Poly(sodium 4-styrene sulfonate) (PSS) e | 200 | 220 | 50 µg/mL | 0.7% |
| Dextran sulfate 500 e | 500 | 470 | 100 µg/mL | 5.2% |
2.2. Protein Function in Crowded Environments
2.2.1. Ordered Monomeric Proteins
2.2.2. Ordered Oligomeric Proteins
2.2.3. Effect of Macromolecular Crowding on Small-Molecule Substrates
2.3. Structural Perturbations of Intrinsically Disordered Proteins in Crowded Milieu
2.4. Effects of Macromolecular Crowding on Shape of Protein Molecules
2.5. Conformational Behavior and Folding of Proteins in Crowded Environment
2.5.1. Effects of Crowded Milieu on Protein Conformational Stability
2.5.2. Altered Conformational Behavior of Proteins in Crowded Environments
2.5.3. Structural Changes Induced in Globular Proteins by Macromolecular Crowding
2.5.4. Protein Folding in Crowded Milieu
2.6. Protein Assembly in a Crowded Environment
2.7. Phase Separation and Compartmentalization in a Crowded Environment
2.8. Pathogenic Protein Aggregation and Fibrillation in a Crowded Environment
2.9. Inequality of Crowding Agents
2.10. Effects of Mixed Macromolecular Crowding Agents
3. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zimmerman, S.B.; Trach, S.O. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of escherichia coli. J. Mol. Biol. 1991, 222, 599–620. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, B.; Ellis, R.J.; Dobson, C.M. Effects of macromolecular crowding on protein folding and aggregation. EMBO J. 1999, 18, 6927–6933. [Google Scholar]
- Rivas, G.; Ferrone, F.; Herzfeld, J. Life in a crowded world. EMBO Rep. 2004, 5, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.J.; Minton, A.P. Cell biology: Join the crowd. Nature 2003, 425, 27–28. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.B.; Minton, A.P. Macromolecular crowding: Biochemical, biophysical, and physiological consequences. Annu. Rev. Biophys. Biomol. Struct. 1993, 22, 27–65. [Google Scholar] [CrossRef] [PubMed]
- Fulton, A.B. How crowded is the cytoplasm? Cell 1982, 30, 345–347. [Google Scholar] [CrossRef] [PubMed]
- Minton, A.P. Influence of excluded volume upon macromolecular structure and associations in “crowded” media. Curr. Opin. Biotechnol. 1997, 8, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.J. Macromolecular crowding: Obvious but underappreciated. Trends Biochem. Sci. 2001, 26, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Minton, A.P. Protein folding: Thickening the broth. Curr. Biol. 2000, 10, R97–R99. [Google Scholar] [CrossRef] [PubMed]
- Lodish, H.; Berk, A.; Zipursky, S.L.; Matsudaira, P.; Baltimore, D.; Darnell, J. Molecular Cell Biology,, 4th ed.; W.H. Freeman: New York, NY, USA, 2000. [Google Scholar]
- Minton, A.P. Implications of macromolecular crowding for protein assembly. Curr. Opin. Struct. Biol. 2000, 10, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Homouz, D.; Perham, M.; Samiotakis, A.; Cheung, M.S.; Wittung-Stafshede, P. Crowded, cell-like environment induces shape changes in aspherical protein. Proc. Natl. Acad. Sci. USA 2008, 105, 11754–11759. [Google Scholar] [CrossRef] [PubMed]
- Minton, A.P. Models for excluded volume interaction between an unfolded protein and rigid macromolecular cosolutes: Macromolecular crowding and protein stability revisited. Biophys. J. 2005, 88, 971–985. [Google Scholar] [CrossRef] [PubMed]
- Minton, A.P. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J. Biol. Chem. 2001, 276, 10577–10580. [Google Scholar] [CrossRef] [PubMed]
- Ralston, G.B. Effects of “crowding” in protein solutions. J. Chem. Edu. 2005, 67, 857–860. [Google Scholar] [CrossRef]
- Eggers, D.K.; Valentine, J.S. Crowding and hydration effects on protein conformation: A study with sol–gel encapsulated proteins. J. Mol. Biol. 2001, 314, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Eggers, D.K.; Valentine, J.S. Molecular confinement influences protein structure and enhances thermal protein stability. Protein Sci. 2001, 10, 250–261. [Google Scholar] [CrossRef] [PubMed]
- Bismuto, E.; Martelli, P.L.; de Maio, A.; Mita, D.G.; Irace, G.; Casadio, R. Effect of molecular confinement on internal enzyme dynamics: Frequency domain fluorometry and molecular dynamics simulation studies. Biopolymers 2002, 67, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Morar, A.S.; Olteanu, A.; Young, G.B.; Pielak, G.J. Solvent-induced collapse of alpha-synuclein and acid-denatured cytochrome c. Protein Sci. 2001, 10, 2195–2199. [Google Scholar] [CrossRef] [PubMed]
- Hatters, D.M.; Minton, A.P.; Howlett, G.J. Macromolecular crowding accelerates amyloid formation by human apolipoprotein c-ii. J. Biol. Chem. 2002, 277, 7824–7830. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Cooper, E.M.; Bower, K.S.; Li, J.; Fink, A.L. Accelerated alpha-synuclein fibrillation in crowded milieu. FEBS Lett. 2002, 515, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Shtilerman, M.D.; Ding, T.T.; Lansbury, P.T., Jr. Molecular crowding accelerates fibrillization of alpha-synuclein: Could an increase in the cytoplasmic protein concentration induce parkinsonʼs disease? Biochemistry 2002, 41, 3855–3860. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Loe, F.; Blocki, A.; Peng, Y.; Raghunath, M. Applying macromolecular crowding to enhance extracellular matrix deposition and its remodeling in vitro for tissue engineering and cell-based therapies. Adv. Drug Deliv. Rev. 2011, 63, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Minton, A.P. Macromolecular crowding and molecular recognition. J. Mol. Recognit. 1993, 6, 211–214. [Google Scholar] [CrossRef] [PubMed]
- Tokuriki, N.; Kinjo, M.; Negi, S.; Hoshino, M.; Goto, Y.; Urabe, I.; Yomo, T. Protein folding by the effects of macromolecular crowding. Protein Sci. 2004, 13, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Lan, E.H.; Dave, B.C.; Fukuto, J.M.; Dunn, B.; Zink, J.I.; Valentine, J.S. Synthesis of sol–gel encapsulated heme proteins with chemical sensing properties. J. Mater. Chem. 1999, 9, 45–53. [Google Scholar] [CrossRef]
- Gottfried, D.S.; Kagan, A.; Hoffman, B.M.; Friedman, J.M. Impeded rotation of a protein in sol–gel matrix. J. Phys. Chem. B 1999, 103, 2803–2807. [Google Scholar] [CrossRef]
- Brennan, J.D. Using fluorescence to investigate proteins entrapped in sol–gel derived materials. Appl. Spec. 1999, 53, 106A–121A. [Google Scholar]
- Dave, B.C.; Dunn, B.; Valentine, J.S.; Zink, J.I. Sol–gel encapsulation methods for biosensors. Anal. Chem. 1994, 66, 1120–1127. [Google Scholar] [CrossRef]
- Gill, I.; Ballestros, A. Bioencapsulation within synthetic polymers (part 1): Sol–gel encapsulated biologicals. Trends Biotechnol. 2000, 18, 282–296. [Google Scholar] [CrossRef] [PubMed]
- Bismuto, E.; Irace, G. The effect of molecular confinement on the conformational dynamics of the native and partly folded state of apomyoglobin. FEBS Lett. 2001, 509, 476–480. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Venable, R.M.; Mackerell, A.D., Jr.; Pastor, R.W. Molecular dynamics studies of polyethylene oxide and polyethylene glycol: Hydrodynamic radius and shape anisotropy. Biophys. J. 2008, 95, 1590–1599. [Google Scholar] [CrossRef] [PubMed]
- Devanand, K.; Selser, J.C. Asymptotic behavior and long-range interactions in aqueous solutions of poly(ethylene oxide). Macromolecules 1991, 24, 5943–5947. [Google Scholar] [CrossRef]
- Uversky, V.N. Use of fast protein size-exclusion liquid chromatography to study the unfolding of proteins which denature through the molten globule. Biochemistry 1993, 32, 13288–13298. [Google Scholar] [CrossRef] [PubMed]
- Linegar, K.L.; Adeniran, A.E.; Kostko, A.F.; Anisimov, M.A. Hydrodynamic radius of polyethylene glycol in solution obtained by dynamic light scattering. Colloid J. 2010, 72, 279–281. [Google Scholar] [CrossRef]
- Zimmerman, S.B.; Pheiffer, B.H. Macromolecular crowding allows blunt-end ligation by DNA ligases from rat liver or escherichia coli. Proc. Natl. Acad. Sci. USA 1983, 80, 5852–5856. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.B.; Harrison, B. Macromolecular crowding accelerates the cohesion of DNA fragments with complementary termini. Nucleic Acids Res. 1985, 13, 2241–2249. [Google Scholar] [CrossRef] [PubMed]
- Harrison, B.; Zimmerman, S.B. Polymer-stimulated ligation: Enhanced ligation of oligo- and polynucleotides by t4 rna ligase in polymer solutions. Nucleic Acids Res. 1984, 12, 8235–8251. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.B.; Harrison, B. Macromolecular crowding increases binding of DNA polymerase to DNA: An adaptive effect. Proc. Natl. Acad. Sci. USA 1987, 84, 1871–1875. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.B.; Trach, S.O. Macromolecular crowding extends the range of conditions under which DNA polymerase is functional. Biochim. Biophys. Acta 1988, 949, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Lavery, P.E.; Kowalczykowski, S.C. Enhancement of RECA protein-promoted DNA strand exchange activity by volume-occupying agents. J. Biol. Chem. 1992, 267, 9307–9314. [Google Scholar] [PubMed]
- Somalinga, B.R.; Roy, R.P. Volume exclusion effect as a driving force for reverse proteolysis. Implications for polypeptide assemblage in a macromolecular crowded milieu. J. Biol. Chem. 2002, 277, 43253–43261. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.M.; Mo, Z.Y.; Wu, L.J.; Chen, J.; Liang, Y. Binding of calcium ions to RAS promotes RAS guanine nucleotide exchange under emulated physiological conditions. Biochim. Biophys. Acta 2008, 1784, 1560–1569. [Google Scholar] [CrossRef] [PubMed]
- Pozdnyakova, I.; Wittung-Stafshede, P. Non-linear effects of macromolecular crowding on enzymatic activity of multi-copper oxidase. Biochim. Biophys. Acta 2010, 1804, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Pastor, I.; Vilaseca, E.; Madurga, S.; Garces, J.L.; Cascante, M.; Mas, F. Effect of crowding by dextrans on the hydrolysis of N-succinyl-l-phenyl-ala-p-nitroanilide catalyzed by alpha-chymotrypsin. J. Phys. Chem. B 2011, 115, 1115–1121. [Google Scholar] [CrossRef] [PubMed]
- Suthar, M.K.; Doharey, P.K.; Verma, A.; Saxena, J.K. Behavior of plasmodium falciparum purine nucleoside phosphorylase in macromolecular crowded environment. Int. J. Biol. Macromol. 2013, 62, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Guo, Z. Effects of macromolecular crowding on the intrinsic catalytic efficiency and structure of enterobactin-specific isochorismate synthase. J. Am. Chem. Soc. 2007, 129, 730–731. [Google Scholar] [CrossRef]
- Akabayov, S.R.; Akabayov, B.; Richardson, C.C.; Wagner, G. Molecular crowding enhanced atpase activity of the rna helicase eif4a correlates with compaction of its quaternary structure and association with eif4g. J. Am. Chem. Soc. 2013, 135, 10040–10047. [Google Scholar] [CrossRef] [PubMed]
- Aoki, K.; Takahashi, K.; Kaizu, K.; Matsuda, M. A quantitative model of ERK map kinase phosphorylation in crowded media. Sci. Rep. 2013, 3, 1541. [Google Scholar] [CrossRef]
- Aoki, K.; Yamada, M.; Kunida, K.; Yasuda, S.; Matsuda, M. Processive phosphorylation of ERK map kinase in mammalian cells. Proc. Natl. Acad. Sci. USA 2011, 108, 12675–12680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asaad, N.; Engberts, J.B. Cytosol-mimetic chemistry: Kinetics of the trypsin-catalyzed hydrolysis of p-nitrophenyl acetate upon addition of polyethylene glycol and N-tert-butyl acetoacetamide. J. Am. Chem. Soc. 2003, 125, 6874–6875. [Google Scholar] [CrossRef] [PubMed]
- Vopel, T.; Makhatadze, G.I. Enzyme activity in the crowded milieu. PLoS One 2012, 7, e39418. [Google Scholar] [PubMed]
- Laurent, T.C. Enzyme reactions in polymer media. Eur. J. Biochem. 1971, 21, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Minton, A.P.; Wilf, J. Effect of macromolecular crowding upon the structure and function of an enzyme: Glyceraldehyde-3-phosphate dehydrogenase. Biochemistry 1981, 20, 4821–4826. [Google Scholar] [CrossRef] [PubMed]
- Rohwer, J.M.; Postma, P.W.; Kholodenko, B.N.; Westerhoff, H.V. Implications of macromolecular crowding for signal transduction and metabolite channeling. Proc. Natl. Acad. Sci. USA 1998, 95, 10547–10552. [Google Scholar] [CrossRef] [PubMed]
- Kholodenko, B.N.; Rohwer, J.M.; Cascante, M.; Westerhoff, H.V. Subtleties in control by metabolic channeling and enzyme organization. Mol. Cell. Biochem. 1998, 184, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Norris, M.G.; Malys, N. What is the true enzyme kinetics in the biological system? An investigation of macromolecular crowding effect upon enzyme kinetics of glucose-6-phosphate dehydrogenase. Biochem. Biophys. Res. Commun. 2011, 405, 388–392. [Google Scholar] [CrossRef] [PubMed]
- Totani, K.; Ihara, Y.; Matsuo, I.; Ito, Y. Effects of macromolecular crowding on glycoprotein processing enzymes. J. Am. Chem. Soc. 2008, 130, 2101–2107. [Google Scholar] [CrossRef] [PubMed]
- Olsen, S.N. Applications of isothermal titration calorimetry to measure enzyme kinetics and activity in complex solutions. Thermochim. Acta 2006, 448, 12–18. [Google Scholar] [CrossRef]
- Balcells, C.; Pastor, I.; Vilaseca, E.; Madurga, S.; Cascante, M.; Mas, F. Macromolecular crowding effect upon in vitro enzyme kinetics: Mixed activation-diffusion control of the oxidation of nadh by pyruvate catalyzed by lactate dehydrogenase. J. Phys. Chem. B 2014, 118, 4062–4068. [Google Scholar] [CrossRef] [PubMed]
- Homchaudhuri, L.; Sarma, N.; Swaminathan, R. Effect of crowding by dextrans and ficolls on the rate of alkaline phosphatase-catalyzed hydrolysis: A size-dependent investigation. Biopolymers 2006, 83, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Richter, K.; Nessling, M.; Lichter, P. Macromolecular crowding and its potential impact on nuclear function. Biochim. Biophys. Acta 2008, 1783, 2100–2107. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.; Hartl, F.U. The effect of macromolecular crowding on chaperonin-mediated protein folding. Proc. Natl. Acad. Sci. USA 1997, 94, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Martin, I.; Celaya, G.; Alfonso, C.; Moro, F.; Rivas, G.; Muga, A. Crowding activates CLPB and enhances its association with dnak for efficient protein aggregate reactivation. Biophys. J. 2014, 106, 2017–2027. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.F.; Jiang, M.; Zheng, S.; Guo, Z. Suppression of linear side products by macromolecular crowding in nonribosomal enterobactin biosynthesis. Org. Lett. 2008, 10, 649–652. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.F.; Jiang, M.; Zheng, S.; Guo, Z. Structural change of the enterobactin synthetase in crowded solution and its relation to crowding-enhanced product specificity in nonribosomal enterobactin biosynthesis. Bioorg. Med. Chem. Lett. 2010, 20, 3855–3858. [Google Scholar] [CrossRef] [PubMed]
- Gehring, A.M.; Mori, I.; Perry, R.D.; Walsh, C.T. The nonribosomal peptide synthetase hmwp2 forms a thiazoline ring during biogenesis of yersiniabactin, an iron-chelating virulence factor of yersinia pestis. Biochemistry 1998, 37, 11637–11650. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, D.N.; Oliveira, L.C.; Kondo, M.Y.; Cezari, M.H.; Szeltner, Z.; Juhasz, T.; Juliano, M.A.; Polgar, L.; Juliano, L.; Gouvea, I.E. Increase of sars-cov 3cl peptidase activity due to macromolecular crowding effects in the milieu composition. Biol. Chem. 2010, 391, 1461–1468. [Google Scholar] [CrossRef] [PubMed]
- Kita, A.; Okajima, K.; Morimoto, Y.; Ikeuchi, M.; Miki, K. Structure of a cyanobacterial bluf protein, tll0078, containing a novel fad-binding blue light sensor domain. J. Mol. Biol. 2005, 349, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jung, A.; Reinstein, J.; Domratcheva, T.; Shoeman, R.L.; Schlichting, I. Crystal structures of the appa bluf domain photoreceptor provide insights into blue light-mediated signal transduction. J. Mol. Biol. 2006, 362, 717–732. [Google Scholar] [CrossRef] [PubMed]
- Okajima, K.; Yoshihara, S.; Fukushima, Y.; Geng, X.; Katayama, M.; Higashi, S.; Watanabe, M.; Sato, S.; Tabata, S.; Shibata, Y.; et al. Biochemical and functional characterization of bluf-type flavin-binding proteins of two species of cyanobacteria. J. Biochem. 2005, 137, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Toyooka, T.; Tanaka, K.; Okajima, K.; Ikeuchi, M.; Tokutomi, S.; Terazima, M. Macromolecular crowding effects on reactions of tepixd (tll0078). Photochem. Photobiol. 2010, 87, 584–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aumiller, W.M., Jr.; Davis, B.W.; Hatzakis, E.; Keating, C.D. Interactions of macromolecular crowding agents and cosolutes with small-molecule substrates: Effect on horseradish peroxidase activity with two different substrates. J. Phys. Chem. B 2014, 118, 10624–10632. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999, 293, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 2000, 41, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Obradovic, Z. The protein trinity—Linking function and disorder. Nat. Biotechnol. 2001, 19, 805–806. [Google Scholar] [CrossRef] [PubMed]
- Dyson, H.J.; Wright, P.E. Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 2002, 12, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Brown, C.J.; Obradovic, Z. Identification and functions of usefully disordered proteins. Adv. Protein Chem. 2002, 62, 25–49. [Google Scholar] [PubMed]
- Dunker, A.K.; Brown, C.J.; Lawson, J.D.; Iakoucheva, L.M.; Obradovic, Z. Intrinsic disorder and protein function. Biochemistry 2002, 41, 6573–6582. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002, 27, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002, 11, 739–756. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. What does it mean to be natively unfolded? Eur. J. Biochem. 2002, 269, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Protein folding revisited. A polypeptide chain at the folding-misfolding-nonfolding cross-roads: Which way to go? Cell Mol. Life Sci. 2003, 60, 1852–1871. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P.; Csermely, P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004, 18, 1169–1175. [Google Scholar] [CrossRef] [PubMed]
- Daughdrill, G.W.; Pielak, G.J.; Uversky, V.N.; Cortese, M.S.; Dunker, A.K. Natively disordered proteins. In Handbook of Protein Folding; Buchner, J., Kiefhaber, T., Eds.; Wiley-VCH Verlag GmbH & Co.: Weinheim, Germany, 2005; pp. 271–353. [Google Scholar]
- Dunker, A.K.; Cortese, M.S.; Romero, P.; Iakoucheva, L.M.; Uversky, V.N. Flexible nets: The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005, 272, 5129–5148. [Google Scholar] [CrossRef] [PubMed]
- Dyson, H.J.; Wright, P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005, 6, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Romero, P.; Uversky, V.N.; Dunker, A.K. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry 2005, 44, 12454–12470. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005, 579, 3346–3354. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P.; Szasz, C.; Buday, L. Structural disorder throws new light on moonlighting. Trends Biochem. Sci. 2005, 30, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Oldfield, C.J.; Dunker, A.K. Showing your id: Intrinsic disorder as an ID for recognition, regulation and cell signaling. J. Mol. Recognit. 2005, 18, 343–384. [Google Scholar] [CrossRef] [PubMed]
- Radivojac, P.; Iakoucheva, L.M.; Oldfield, C.J.; Obradovic, Z.; Uversky, V.N.; Dunker, A.K. Intrinsic disorder and functional proteomics. Biophys. J. 2007, 92, 1439–1456. [Google Scholar] [CrossRef] [PubMed]
- Vucetic, S.; Xie, H.; Iakoucheva, L.M.; Oldfield, C.J.; Dunker, A.K.; Obradovic, Z.; Uversky, V.N. Functional anthology of intrinsic disorder. 2. Cellular components, domains, technical terms, developmental processes, and coding sequence diversities correlated with long disordered regions. J. Proteome Res. 2007, 6, 1899–1916. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Vucetic, S.; Iakoucheva, L.M.; Oldfield, C.J.; Dunker, A.K.; Uversky, V.N.; Obradovic, Z. Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J. Proteome Res. 2007, 6, 1882–1898. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Vucetic, S.; Iakoucheva, L.M.; Oldfield, C.J.; Dunker, A.K.; Obradovic, Z.; Uversky, V.N. Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J. Proteome Res. 2007, 6, 1917–1932. [Google Scholar] [CrossRef] [PubMed]
- Cortese, M.S.; Uversky, V.N.; Dunker, A.K. Intrinsic disorder in scaffold proteins: Getting more from less. Prog. Biophys. Mol. Biol. 2008, 98, 85–106. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Silman, I.; Uversky, V.N.; Sussman, J.L. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008, 18, 756–764. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Uversky, V.N. Signal transduction via unstructured protein conduits. Nat. Chem. Biol. 2008, 4, 229–230. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Oldfield, C.J.; Meng, J.; Romero, P.; Yang, J.Y.; Chen, J.W.; Vacic, V.; Obradovic, Z.; Uversky, V.N. The unfoldomics decade: An update on intrinsically disordered proteins. BMC Genomics 2008, 9, S1. [Google Scholar] [CrossRef] [PubMed]
- Oldfield, C.J.; Meng, J.; Yang, J.Y.; Yang, M.Q.; Uversky, V.N.; Dunker, A.K. Flexible nets: Disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics 2008, 9, S1. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.B.; Gibson, T.J. A careful disorderliness in the proteome: Sites for interaction and targets for future therapies. FEBS Lett. 2008, 582, 1271–1275. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P.; Fuxreiter, M. Fuzzy complexes: Polymorphism and structural disorder in protein–protein interactions. Trends Biochem. Sci. 2008, 33, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Dunker, A.K. Biochemistry. Controlled chaos. Science 2008, 322, 1340–1341. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Dunker, A.K. Understanding protein non-folding. Biochim. Biophys. Acta 2010, 1804, 1231–1264. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P.; Fuxreiter, M.; Oldfield, C.J.; Simon, I.; Dunker, A.K.; Uversky, V.N. Close encounters of the third kind: Disordered domains and the interactions of proteins. Bioessays 2009, 31, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Linking folding and binding. Curr. Opin. Struct. Biol. 2009, 19, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Obradovic, Z.; Romero, P.; Garner, E.C.; Brown, C.J. Intrinsic protein disorder in complete genomes. Genome Inform. Ser. Workshop Genome Inform. 2000, 11, 161–171. [Google Scholar] [PubMed]
- Uversky, V.N. The mysterious unfoldome: Structureless, underappreciated, yet vital part of any given proteome. J. Biomed. Biotechnol. 2010, 2010, 568068. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.J.; Sodhi, J.S.; McGuffin, L.J.; Buxton, B.F.; Jones, D.T. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004, 337, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Iakoucheva, L.M.; Brown, C.J.; Lawson, J.D.; Obradovic, Z.; Dunker, A.K. Intrinsic disorder in cell-signaling and cancer-associated proteins. J. Mol. Biol. 2002, 323, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Schulz, G.E. Nucleotide binding proteins. In Molecular Mechanism of Biological Recognition; Balaban, M., Ed.; Elsevier/North-Holland Biomedical Press: New York, NY, USA, 1979; pp. 79–94. [Google Scholar]
- Pontius, B.W. Close encounters: Why unstructured, polymeric domains can increase rates of specific macromolecular association. Trends Biochem. Sci. 1993, 18, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Spolar, R.S.; Record, M.T., Jr. Coupling of local folding to site-specific binding of proteins to DNA. Science 1994, 263, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, R.; Vajda, S.; DeLisi, C. Flexible docking and design. Annu. Rev. Biophys. Biomol. Struct. 1995, 24, 677–700. [Google Scholar] [CrossRef] [PubMed]
- Plaxco, K.W.; Gross, M. Cell biology. The importance of being unfolded. Nature 1997, 386, 657–659. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.M.; Xu, J.; Clarkson, B.K.; Martinez-Yamout, M.A.; Dyson, H.J.; Case, D.A.; Gottesfeld, J.M.; Wright, P.E. Induced fit and “lock and key” recognition of 5S RNA by zinc fingers of transcription factor iiia. J. Mol. Biol. 2006, 357, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Sugase, K.; Dyson, H.J.; Wright, P.E. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 2007, 447, 1021–1025. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Oldfield, C.J.; Dunker, A.K. Intrinsically disordered proteins in human diseases: Introducing the d2 concept. Annu. Rev. Biophys. 2008, 37, 215–246. [Google Scholar] [CrossRef] [PubMed]
- Flaugh, S.L.; Lumb, K.J. Effects of macromolecular crowding on the intrinsically disordered proteins c-fos and p27(kip1). Biomacromolecules 2001, 2, 538–540. [Google Scholar] [CrossRef] [PubMed]
- Campbell, K.M.; Terrell, A.R.; Laybourn, P.J.; Lumb, K.J. Intrinsic structural disorder of the C-terminal activation domain from the BZIP transcription factor fos. Biochemistry 2000, 39, 2708–2713. [Google Scholar] [CrossRef] [PubMed]
- Bienkiewicz, E.A.; Adkins, J.N.; Lumb, K.J. Functional consequences of preorganized helical structure in the intrinsically disordered cell-cycle inhibitor p27(kip1). Biochemistry 2002, 41, 752–759. [Google Scholar] [CrossRef] [PubMed]
- Munishkina, L.A.; Cooper, E.M.; Uversky, V.N.; Fink, A.L. The effect of macromolecular crowding on protein aggregation and amyloid fibril formation. J. Mol. Recognit. 2004, 17, 456–464. [Google Scholar] [CrossRef] [PubMed]
- McNulty, B.C.; Young, G.B.; Pielak, G.J. Macromolecular crowding in the Escherichia coli periplasm maintains alpha-synuclein disorder. J. Mol. Biol. 2006, 355, 893–897. [Google Scholar] [CrossRef] [PubMed]
- Mouillon, J.M.; Eriksson, S.K.; Harryson, P. Mimicking the plant cell interior under water stress by macromolecular crowding: Disordered dehydrin proteins are highly resistant to structural collapse. Plant Physiol. 2008, 148, 1925–1937. [Google Scholar] [CrossRef] [PubMed]
- Dedmon, M.M.; Patel, C.N.; Young, G.B.; Pielak, G.J. Flgm gains structure in living cells. Proc. Natl. Acad. Sci. USA 2002, 99, 12681–12684. [Google Scholar] [CrossRef] [PubMed]
- Daughdrill, G.W.; Hanely, L.J.; Dahlquist, F.W. The C-terminal half of the anti-sigma factor flgm contains a dynamic equilibrium solution structure favoring helical conformations. Biochemistry 1998, 37, 1076–1082. [Google Scholar] [CrossRef] [PubMed]
- Daughdrill, G.W.; Chadsey, M.S.; Karlinsey, J.E.; Hughes, K.T.; Dahlquist, F.W. The C-terminal half of the anti-sigma factor, flgm, becomes structured when bound to its target, sigma 28. Nat. Struct. Biol. 1997, 4, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Roque, A.; Ponte, I.; Suau, P. Macromolecular crowding induces a molten globule state in the C-terminal domain of histone h1. Biophys. J. 2007, 93, 2170–2177. [Google Scholar] [CrossRef] [PubMed]
- Aden, J.; Wittung-Stafshede, P. Folding of an unfolded protein by macromolecular crowding in vitro. Biochemistry 2014, 53, 2271–2277. [Google Scholar] [CrossRef] [PubMed]
- Johansen, D.; Jeffries, C.M.; Hammouda, B.; Trewhella, J.; Goldenberg, D.P. Effects of macromolecular crowding on an intrinsically disordered protein characterized by small-angle neutron scattering with contrast matching. Biophys. J. 2011, 100, 1120–1128. [Google Scholar] [CrossRef] [PubMed]
- Goldenberg, D.P.; Argyle, B. Minimal effects of macromolecular crowding on an intrinsically disordered protein: A small-angle neutron scattering study. Biophys. J. 2014, 106, 905–914. [Google Scholar] [CrossRef] [PubMed]
- Soranno, A.; Koenig, I.; Borgia, M.B.; Hofmann, H.; Zosel, F.; Nettels, D.; Schuler, B. Single-molecule spectroscopy reveals polymer effects of disordered proteins in crowded environments. Proc. Natl. Acad. Sci. USA 2014, 111, 4874–4879. [Google Scholar] [CrossRef] [PubMed]
- Sotomayor-Perez, A.C.; Ladant, D.; Chenal, A. Calcium-induced folding of intrinsically disordered repeat-in-toxin (rtx) motifs via changes of protein charges and oligomerization states. J. Biol. Chem. 2011, 286, 16997–17004. [Google Scholar] [CrossRef] [PubMed]
- Sotomayor-Perez, A.C.; Subrini, O.; Hessel, A.; Ladant, D.; Chenal, A. Molecular crowding stabilizes both the intrinsically disordered calcium-free state and the folded calcium-bound state of a repeat in toxin (rtx) protein. J. Am. Chem. Soc. 2013, 135, 11929–11934. [Google Scholar] [CrossRef] [PubMed]
- Cino, E.A.; Karttunen, M.; Choy, W.Y. Effects of molecular crowding on the dynamics of intrinsically disordered proteins. PLoS One 2012, 7, e49876. [Google Scholar] [CrossRef] [PubMed]
- Eicken, C.; Sharma, V.; Klabunde, T.; Lawrenz, M.B.; Hardham, J.M.; Norris, S.J.; Sacchettini, J.C. Crystal structure of lyme disease variable surface antigen vlse of borrelia burgdorferi. J. Biol. Chem. 2002, 277, 21691–21696. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.; Wittung-Stafshede, P. The largest protein observed to fold by two-state kinetic mechanism does not obey contact-order correlation. J. Am. Chem. Soc. 2003, 125, 9606–9607. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.; Guidry, J.; Wittung-Stafshede, P. Characterization of surface antigen from lyme disease spirochete borrelia burgdorferi. Biochem. Biophys. Res. Commun. 2001, 289, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Homouz, D.; Sanabria, H.; Waxham, M.N.; Cheung, M.S. Modulation of calmodulin plasticity by the effect of macromolecular crowding. J. Mol. Biol. 2009, 391, 933–943. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liang, K.C.; Czader, A.; Waxham, M.N.; Cheung, M.S. The effect of macromolecular crowding, ionic strength and calcium binding on calmodulin dynamics. PLoS Comput. Biol. 2011, 7, e1002114. [Google Scholar] [CrossRef] [PubMed]
- Dhar, A.; Samiotakis, A.; Ebbinghaus, S.; Nienhaus, L.; Homouz, D.; Gruebele, M.; Cheung, M.S. Structure, function, and folding of phosphoglycerate kinase are strongly perturbed by macromolecular crowding. Proc. Natl. Acad. Sci. USA 2010, 107, 17586–17591. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.M.; Chyan, C.L.; Zhou, H.X.; Chung, T.Y.; Peng, H.; Ping, G.; Yang, G. The effects of macromolecular crowding on the mechanical stability of protein molecules. Protein Sci. 2008, 17, 2156–2166. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, A.; Wang, Q.; Samiotakis, A.; Cheung, M.S.; Wittung-Stafshede, P. Factors defining effects of macromolecular crowding on protein stability: An in vitro/in silico case study using cytochrome c. Biochemistry 2010, 49, 6519–6530. [Google Scholar] [CrossRef] [PubMed]
- Mikaelsson, T.; Aden, J.; Wittung-Stafshede, P.; Johansson, L.B. Macromolecular crowding effects on two homologs of ribosomal protein s16: Protein-dependent structural changes and local interactions. Biophys. J. 2014, 107, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Engel, R.; Westphal, A.H.; Huberts, D.H.; Nabuurs, S.M.; Lindhoud, S.; Visser, A.J.; van Mierlo, C.P. Macromolecular crowding compacts unfolded apoflavodoxin and causes severe aggregation of the off-pathway intermediate during apoflavodoxin folding. J. Biol. Chem. 2008, 283, 27383–27394. [Google Scholar] [CrossRef] [PubMed]
- Mittal, S.; Singh, L.R. Denatured state structural property determines protein stabilization by macromolecular crowding: A thermodynamic and structural approach. PLoS One 2013, 8, e78936. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; He, H.; Li, S. Effect of ficoll 70 on thermal stability and structure of creatine kinase. Biochemistry 2010, 75, 648–654. [Google Scholar] [PubMed]
- Aguilar, X.; C, F.W.; Sparrman, T.; Wolf-Watz, M.; Wittung-Stafshede, P. Macromolecular crowding extended to a heptameric system: The co-chaperonin protein 10. Biochemistry 2011, 50, 3034–3044. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.Q.; Liu, H.J.; Li, C.; Luan, Y.S.; Yang, J.M.; Wang, Y.L. Inactivation of recombinant human brain-type creatine kinase during denaturation by guanidine hydrochloride in a macromolecular crowding system. Appl. Biochem. Biotechnol. 2013, 169, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Winter, R. Effect of molecular crowding on the temperature-pressure stability diagram of ribonuclease a. Chemphyschem 2013, 14, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Erlkamp, M.; Grobelny, S.; Winter, R. Crowding effects on the temperature and pressure dependent structure, stability and folding kinetics of staphylococcal nuclease. Phys. Chem. Chem. Phys. 2014, 16, 5965–5976. [Google Scholar] [CrossRef] [PubMed]
- Pollard, T.D.; Blanchoin, L.; Mullins, R.D. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 545–576. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, S.S.; Kerwar, G. Intrinsic fluorescence of actin. Biochemistry 1972, 11, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Strzelecka-Golaszewska, H.; Nagy, B.; Gergely, J. Changes in conformation and nucleotide binding of ca, mn, or mgg-actin upon removal of the bound divalent cation. Studies of ultraviolet difference spectra and optical rotation. Arch. Biochem. Biophys. 1974, 161, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Strzelecka-Golaszewska, H.; Venyaminov, S.; Zmorzynski, S.; Mossakowska, M. Effects of various amino acid replacements on the conformational stability of g-actin. Eur. J. Biochem. 1985, 147, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Tellam, R.L.; Sculley, M.J.; Nichol, L.W.; Wills, P.R. The influence of poly(ethylene glycol) 6000 on the properties of skeletal-muscle actin. Biochem. J. 1983, 213, 651–659. [Google Scholar] [PubMed]
- Qu, Y.; Bolen, D.W. Efficacy of macromolecular crowding in forcing proteins to fold. Biophys. Chem. 2002, 101–102, 155–165. [Google Scholar]
- Sasahara, K.; McPhie, P.; Minton, A.P. Effect of dextran on protein stability and conformation attributed to macromolecular crowding. J. Mol. Biol. 2003, 326, 1227–1237. [Google Scholar] [CrossRef] [PubMed]
- McPhie, P.; Ni, Y.S.; Minton, A.P. Macromolecular crowding stabilizes the molten globule form of apomyoglobin with respect to both cold and heat unfolding. J. Mol. Biol. 2006, 361, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Stagg, L.; Zhang, S.Q.; Cheung, M.S.; Wittung-Stafshede, P. Molecular crowding enhances native structure and stability of alpha/beta protein flavodoxin. Proc. Natl. Acad. Sci. USA 2007, 104, 18976–18981. [Google Scholar] [CrossRef] [PubMed]
- Perham, M.; Stagg, L.; Wittung-Stafshede, P. Macromolecular crowding increases structural content of folded proteins. FEBS Lett. 2007, 581, 5065–5069. [Google Scholar] [CrossRef] [PubMed]
- Charlton, L.M.; Barnes, C.O.; Li, C.; Orans, J.; Young, G.B.; Pielak, G.J. Residue-level interrogation of macromolecular crowding effects on protein stability. J. Am. Chem. Soc. 2008, 130, 6826–6830. [Google Scholar] [CrossRef] [PubMed]
- Miklos, A.C.; Li, C.; Pielak, G.J. Using nmr-detected backbone amide 1h exchange to assess macromolecular crowding effects on globular-protein stability. Methods Enzymol. 2009, 466, 1–18. [Google Scholar] [PubMed]
- Ai, X.; Zhou, Z.; Bai, Y.; Choy, W.Y. 15n NMR spin relaxation dispersion study of the molecular crowding effects on protein folding under native conditions. J. Am. Chem. Soc. 2006, 128, 3916–3917. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, S.; Wang, C. Effects of macromolecular crowding on the refolding of glucose-6-phosphate dehydrogenase and protein disulfide isomerase. J. Biol. Chem. 2001, 276, 34396–34401. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, B.; Wain, R.; Dobson, C.M.; Ellis, R.J. Macromolecular crowding perturbs protein refolding kinetics: Implications for folding inside the cell. EMBO J. 2000, 19, 3870–3875. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.X. Loops, linkages, rings, catenanes, cages, and crowders: Entropy-based strategies for stabilizing proteins. Acc. Chem. Res. 2004, 37, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Cheung, M.S.; Klimov, D.; Thirumalai, D. Molecular crowding enhances native state stability and refolding rates of globular proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 4753–4758. [Google Scholar] [CrossRef] [PubMed]
- Mikaelsson, T.; Aden, J.; Johansson, L.B.; Wittung-Stafshede, P. Direct observation of protein unfolded state compaction in the presence of macromolecular crowding. Biophys. J. 2013, 104, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Ladurner, A.G.; Fersht, A.R. Upper limit of the time scale for diffusion and chain collapse in chymotrypsin inhibitor 2. Nat. Struct. Biol. 1999, 6, 28–31. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.R.; Liang, Y.; Du, F.; Zhou, Z.; Chen, J. Mixed macromolecular crowding accelerates the oxidative refolding of reduced, denatured lysozyme: Implications for protein folding in intracellular environments. J. Biol. Chem. 2004, 279, 55109–55116. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Waegele, M.M.; Chowdhury, P.; Guo, L.; Gai, F. Effect of macromolecular crowding on protein folding dynamics at the secondary structure level. J. Mol. Biol. 2009, 393, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Galan, A.; Sot, B.; Llorca, O.; Carrascosa, J.L.; Valpuesta, J.M.; Muga, A. Excluded volume effects on the refolding and assembly of an oligomeric protein. Groel, a case study. J. Biol. Chem. 2001, 276, 957–964. [Google Scholar] [CrossRef] [PubMed]
- Martin, J. Requirement for groel/groes-dependent protein folding under nonpermissive conditions of macromolecular crowding. Biochemistry 2002, 41, 5050–5055. [Google Scholar] [CrossRef] [PubMed]
- Ren, G.; Lin, Z.; Tsou, C.L.; Wang, C.C. Effects of macromolecular crowding on the unfolding and the refolding of d-glyceraldehyde-3-phosophospate dehydrogenase. J. Protein Chem. 2003, 22, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Gierasch, L.M. Macromolecular crowding remodels the energy landscape of a protein by favoring a more compact unfolded state. J. Am. Chem. Soc. 2010, 132, 10445–10452. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, A.; Wittung-Stafshede, P. Quantification of excluded volume effects on the folding landscape of pseudomonas aeruginosa apoazurin in vitro. Biophys. J. 2013, 105, 1689–1699. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.; Christiansen, A.; Wang, Q.; Cheung, M.S.; Kliger, D.S.; Wittung-Stafshede, P. Effects of macromolecular crowding on burst phase kinetics of cytochrome c folding. Biochemistry 2012, 51, 9836–9845. [Google Scholar] [CrossRef] [PubMed]
- Stagg, L.; Christiansen, A.; Wittung-Stafshede, P. Macromolecular crowding tunes folding landscape of parallel alpha/beta protein, apoflavodoxin. J. Am. Chem. Soc. 2011, 133, 646–648. [Google Scholar] [CrossRef] [PubMed]
- Stagg, L.; Samiotakis, A.; Homouz, D.; Cheung, M.S.; Wittung-Stafshede, P. Residue-specific analysis of frustration in the folding landscape of repeat beta/alpha protein apoflavodoxin. J. Mol. Biol. 2010, 396, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Homouz, D.; Stagg, L.; Wittung-Stafshede, P.; Cheung, M.S. Macromolecular crowding modulates folding mechanism of alpha/beta protein apoflavodoxin. Biophys. J. 2009, 96, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, A.P.; Wang, Y.; Tadeo, X.; Millet, O.; Pielak, G.J. Macromolecular crowding fails to fold a globular protein in cells. J. Am. Chem. Soc. 2011, 133, 8082–8085. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Chai, Q.; Zhong, M.; Wei, Y. Effect of crowding by ficolls on ompa and ompt refolding and membrane insertion. Protein Sci. 2013, 22, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.B.; Trach, S.O. Effects of macromolecular crowding on the association of E. coli ribosomal particles. Nucleic Acids Res. 1988, 16, 6309–6326. [Google Scholar] [CrossRef] [PubMed]
- Agius, L. Substrate modulation of aldolase b binding in hepatocytes. Biochem. J. 1996, 315, 651–658. [Google Scholar] [PubMed]
- Azoulay-Zohar, H.; Aflalo, C. Binding of rat brain hexokinase to recombinant yeast mitochondria: Identification of necessary molecular determinants. J. Bioenerg. Biomembr. 1999, 31, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Azoulay-Zohar, H.; Aflalo, C. Binding of rat brain hexokinase to recombinant yeast mitochondria: Identification of necessary physico-chemical determinants. Eur. J. Biochem. 2000, 267, 2973–2980. [Google Scholar] [CrossRef] [PubMed]
- Naddaf, L.; Sayyed-Ahmad, A. Intracellular crowding effects on the self-association of the bacterial cell division protein ftsz. Arch. Biochem. Biophys. 2014, 564, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Rivas, G.; Fernandez, J.A.; Minton, A.P. Direct observation of the enhancement of noncooperative protein self-assembly by macromolecular crowding: Indefinite linear self-association of bacterial cell division protein FTSZ. Proc. Natl. Acad. Sci. USA 2001, 98, 3150–3155. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, J.M.; Jimenez, M.; Velez, M.; Mingorance, J.; Andreu, J.M.; Vicente, M.; Rivas, G. Essential cell division protein ftsz assembles into one monomer-thick ribbons under conditions resembling the crowded intracellular environment. J. Biol. Chem. 2003, 278, 37664–37671. [Google Scholar] [CrossRef] [PubMed]
- Dannies, P. Manipulating the reversible aggregation of protein hormones in secretory granules: Potential impact on biopharmaceutical development. BioDrugs 2003, 17, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Del Alamo, M.; Rivas, G.; Mateu, M.G. Effect of macromolecular crowding agents on human immunodeficiency virus type 1 capsid protein assembly in vitro. J. Virol. 2005, 79, 14271–14281. [Google Scholar] [CrossRef] [PubMed]
- Frenkiel-Krispin, D.; Wolf, S.G.; Albeck, S.; Unger, T.; Peleg, Y.; Jacobovitch, J.; Michael, Y.; Daube, S.; Sharon, M.; Robinson, C.V.; et al. Plant transformation by agrobacterium tumefaciens: Modulation of single-stranded DNA-vire2 complex assembly by vire1. J. Biol. Chem. 2007, 282, 3458–3464. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Lopez, T.; Davila-Fajardo, C.; Blaesing, F.; Lillo, M.P.; Giraldo, R. Early events in the binding of the pps10 replication protein repa to single iteron and operator DNA sequences. J. Mol. Biol. 2006, 364, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.L.; Liao, J.M.; Chen, J.; Liang, Y. Macromolecular crowding enhances the binding of superoxide dismutase to xanthine oxidase: Implications for protein-protein interactions in intracellular environments. Int. J. Biochem. Cell Biol. 2006, 38, 1986–1994. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R. Self-association of polynucleosome chains by macromolecular crowding. Eur. Biophys. J. 2008, 37, 1059–1064. [Google Scholar] [CrossRef] [PubMed]
- Alcorlo, M.; Jimenez, M.; Ortega, A.; Hermoso, J.M.; Salas, M.; Minton, A.P.; Rivas, G. Analytical ultracentrifugation studies of phage phi29 protein p6 binding to DNA. J. Mol. Biol. 2009, 385, 1616–1629. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Yethiraj, A. Crowding effects on association reactions at membranes. Biophys. J. 2010, 98, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Jin, R.; Li, J.; Luo, K.; Huang, T.; Wu, D.; Wang, W.; Chen, R.; Xiao, G. Macromolecular crowding converts the human recombinant PRPC to the soluble neurotoxic beta-oligomers. FASEB J. 2010, 24, 3536–3543. [Google Scholar] [CrossRef] [PubMed]
- Snoussi, K.; Halle, B. Protein self-association induced by macromolecular crowding: A quantitative analysis by magnetic relaxation dispersion. Biophys. J. 2005, 88, 2855–2866. [Google Scholar] [CrossRef] [PubMed]
- Wilf, J.; Minton, A.P. Evidence for protein self-association induced by excluded volume. Myoglobin in the presence of globular proteins. Biochim. Biophys. Acta 1981, 670, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Bookchin, R.M.; Balazs, T.; Wang, Z.; Josephs, R.; Lew, V.L. Polymer structure and solubility of deoxyhemoglobin s in the presence of high concentrations of volume-excluding 70-kda dextran. Effects of non-S hemoglobins and inhibitors. J. Biol. Chem. 1999, 274, 6689–6697. [Google Scholar] [CrossRef] [PubMed]
- Lindner, R.A.; Ralston, G.B. Macromolecular crowding: Effects on actin polymerisation. Biophys. Chem. 1997, 66, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.C.; Best, R.B.; Mittal, J. Macromolecular crowding effects on protein-protein binding affinity and specificity. J. Chem. Phys. 2010, 133, 205101. [Google Scholar] [CrossRef] [PubMed]
- Phillip, Y.; Sherman, E.; Haran, G.; Schreiber, G. Common crowding agents have only a small effect on protein-protein interactions. Biophys. J. 2009, 97, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Petrovska, I.; Nuske, E.; Munder, M.C.; Kulasegaran, G.; Malinovska, L.; Kroschwald, S.; Richter, D.; Fahmy, K.; Gibson, K.; Verbavatz, J.M.; et al. Filament formation by metabolic enzymes is a specific adaptation to an advanced state of cellular starvation. eLife 2014. [CrossRef]
- Jarvis, T.C.; Ring, D.M.; Daube, S.S.; von Hippel, P.H. “Macromolecular crowding”: Thermodynamic consequences for protein–protein interactions within the t4 DNA replication complex. J. Biol. Chem. 1990, 265, 15160–15167. [Google Scholar] [PubMed]
- Reddy, M.K.; Weitzel, S.E.; von Hippel, P.H. Assembly of a functional replication complex without atp hydrolysis: A direct interaction of bacteriophage t4 gp45 with t4 DNA polymerase. Proc. Natl. Acad. Sci. USA 1993, 90, 3211–3215. [Google Scholar] [CrossRef] [PubMed]
- Sanders, G.M.; Kassavetis, G.A.; Geiduschek, E.P. Use of a macromolecular crowding agent to dissect interactions and define functions in transcriptional activation by a DNA-tracking protein: Bacteriophage t4 gene 45 protein and late transcription. Proc. Natl. Acad. Sci. USA 1994, 91, 7703–7707. [Google Scholar] [CrossRef] [PubMed]
- Reddy, M.K.; Weitzel, S.E.; Daube, S.S.; Jarvis, T.C.; von Hippel, P.H. Using macromolecular crowding agents to identify weak interactions within DNA replication complexes. Methods Enzymol. 1995, 262, 466–476. [Google Scholar] [PubMed]
- Herendeen, D.R.; Kassavetis, G.A.; Barry, J.; Alberts, B.M.; Geiduschek, E.P. Enhancement of bacteriophage t4 late transcription by components of the t4 DNA replication apparatus. Science 1989, 245, 952–958. [Google Scholar] [CrossRef] [PubMed]
- Herendeen, D.R.; Williams, K.P.; Kassavetis, G.A.; Geiduschek, E.P. An RNA polymerase-binding protein that is required for communication between an enhancer and a promoter. Science 1990, 248, 573–578. [Google Scholar] [CrossRef] [PubMed]
- Herendeen, D.R.; Kassavetis, G.A.; Geiduschek, E.P. A transcriptional enhancer whose function imposes a requirement that proteins track along DNA. Science 1992, 256, 1298–1303. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.J.; Sanders, G.M.; OʼDonnell, M.; Geiduschek, E.P. Dynamics of DNA-tracking by two sliding-clamp proteins. EMBO J. 1996, 15, 4414–4422. [Google Scholar] [PubMed]
- Suzuki, A.; Yamazaki, M.; Ito, T. Osmoelastic coupling in biological structures: Formation of parallel bundles of actin filaments in a crystalline-like structure caused by osmotic stress. Biochemistry 1989, 28, 6513–6518. [Google Scholar] [CrossRef] [PubMed]
- Shearwin, K.; Nanhua, C.; Masters, C. The influence of molecular crowding on the binding of glycolytic enzymes to cytoskeletal structure. Biochem. Int. 1989, 19, 723–729. [Google Scholar] [PubMed]
- Shearwin, K.; Nanhua, C.; Masters, C. Interactions between glycolytic enzymes and cytoskeletal structure—The influence of ionic strength and molecular crowding. Biochem. Int. 1990, 21, 53–60. [Google Scholar] [PubMed]
- Cuneo, P.; Magri, E.; Verzola, A.; Grazi, E. “macromolecular crowding” is a primary factor in the organization of the cytoskeleton. Biochem. J. 1992, 281, 507–512. [Google Scholar] [PubMed]
- Grazi, E. Cytoskeleton, motile structures and macromolecular crowding. Adv. Exp. Med. Biol. 1994, 358, 123–130. [Google Scholar] [PubMed]
- Dewavrin, J.Y.; Hamzavi, N.; Shim, V.P.; Raghunath, M. Tuning the architecture of three-dimensional collagen hydrogels by physiological macromolecular crowding. Acta Biomater. 2014, 10, 4351–4359. [Google Scholar] [CrossRef] [PubMed]
- Minton, A.P.; Colclasure, G.C.; Parker, J.C. Model for the role of macromolecular crowding in regulation of cellular volume. Proc. Natl. Acad. Sci. USA 1992, 89, 10504–10506. [Google Scholar] [CrossRef] [PubMed]
- Rosania, G.R.; Swanson, J.A. Effects of macromolecular crowding on nuclear size. Exp. Cell Res. 1995, 218, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Rincon, V.; Bocanegra, R.; Rodriguez-Huete, A.; Rivas, G.; Mateu, M.G. Effects of macromolecular crowding on the inhibition of virus assembly and virus-cell receptor recognition. Biophys. J. 2011, 100, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Brangwynne, C.P. Phase transitions and size scaling of membrane-less organelles. J. Cell Biol. 2013, 203, 875–881. [Google Scholar] [CrossRef] [PubMed]
- Maul, G.G.; Negorev, D.; Bell, P.; Ishov, A.M. Review: Properties and assembly mechanisms of nd10, pml bodies, or pods. J. Struct. Biol. 2000, 129, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Huang, S. Review: Perinucleolar structures. J. Struct. Biol. 2000, 129, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Fox, A.H.; Lam, Y.W.; Leung, A.K.; Lyon, C.E.; Andersen, J.; Mann, M.; Lamond, A.I. Paraspeckles: A novel nuclear domain. Curr. Biol. 2002, 12, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Lamond, A.I.; Spector, D.L. Nuclear speckles: A model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 2003, 4, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Shav-Tal, Y.; Blechman, J.; Darzacq, X.; Montagna, C.; Dye, B.T.; Patton, J.G.; Singer, R.H.; Zipori, D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol. Biol. Cell 2005, 16, 2395–2413. [Google Scholar] [CrossRef] [PubMed]
- Decker, C.J.; Teixeira, D.; Parker, R. Edc3p and a glutamine/asparagine-rich domain of lsm4p function in processing body assembly in saccharomyces cerevisiae. J. Cell Biol. 2007, 179, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Brangwynne, C.P.; Eckmann, C.R.; Courson, D.S.; Rybarska, A.; Hoege, C.; Gharakhani, J.; Julicher, F.; Hyman, A.A. Germline p granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009, 324, 1729–1732. [Google Scholar] [CrossRef] [PubMed]
- Chuma, S.; Hosokawa, M.; Tanaka, T.; Nakatsuji, N. Ultrastructural characterization of spermatogenesis and its evolutionary conservation in the germline: Germinal granules in mammals. Mol. Cell. Endocrinol. 2009, 306, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Strzelecka, M.; Trowitzsch, S.; Weber, G.; Luhrmann, R.; Oates, A.C.; Neugebauer, K.M. Coilin-dependent snrnp assembly is essential for zebrafish embryogenesis. Nat. Struct. Mol. Biol. 2010, 17, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Decker, M.; Jaensch, S.; Pozniakovsky, A.; Zinke, A.; OʼConnell, K.F.; Zachariae, W.; Myers, E.; Hyman, A.A. Limiting amounts of centrosome material set centrosome size in C. elegans embryos. Curr. Biol. 2011, 21, 1259–1267. [Google Scholar] [CrossRef] [PubMed]
- Wippich, F.; Bodenmiller, B.; Trajkovska, M.G.; Wanka, S.; Aebersold, R.; Pelkmans, L. Dual specificity kinase dyrk3 couples stress granule condensation/dissolution to mtorc1 signaling. Cell 2013, 152, 791–805. [Google Scholar] [CrossRef] [PubMed]
- Phair, R.D.; Misteli, T. High mobility of proteins in the mammalian cell nucleus. Nature 2000, 404, 604–609. [Google Scholar] [CrossRef] [PubMed]
- Pederson, T. Protein mobility within the nucleus--what are the right moves? Cell 2001, 104, 635–638. [Google Scholar] [CrossRef] [PubMed]
- Handwerger, K.E.; Cordero, J.A.; Gall, J.G. Cajal bodies, nucleoli, and speckles in the xenopus oocyte nucleus have a low-density, sponge-like structure. Mol. Biol. Cell 2005, 16, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Updike, D.L.; Hachey, S.J.; Kreher, J.; Strome, S. P granules extend the nuclear pore complex environment in the C. elegans germ line. J. Cell Biol. 2011, 192, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Brangwynne, C.P.; Mitchison, T.J.; Hyman, A.A. Active liquid-like behavior of nucleoli determines their size and shape in xenopus laevis oocytes. Proc. Natl. Acad. Sci. USA 2011, 108, 4334–4339. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Banjade, S.; Cheng, H.C.; Kim, S.; Chen, B.; Guo, L.; Llaguno, M.; Hollingsworth, J.V.; King, D.S.; Banani, S.F.; et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 2012, 483, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, S.; Snaidero, N.; Pahler, G.; Frey, S.; Sanchez, P.; Zweckstetter, M.; Janshoff, A.; Schneider, A.; Weil, M.T.; Schaap, I.A.; et al. Myelin membrane assembly is driven by a phase transition of myelin basic proteins into a cohesive protein meshwork. PLoS Biol. 2013, 11, e1001577. [Google Scholar] [CrossRef] [PubMed]
- Feric, M.; Brangwynne, C.P. A nuclear f-actin scaffold stabilizes ribonucleoprotein droplets against gravity in large cells. Nat. Cell Biol. 2013, 15, 1253–1259. [Google Scholar] [CrossRef] [PubMed]
- Keating, C.D. Aqueous phase separation as a possible route to compartmentalization of biological molecules. Acc. Chem. Res. 2012, 45, 2114–2124. [Google Scholar] [CrossRef] [PubMed]
- Walter, H.; Brooks, D.E. Phase separation in cytoplasm, due to macromolecular crowding, is the basis for microcompartmentation. FEBS Lett. 1995, 361, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Walter, H. Consequences of phase separation in cytoplasm. Int. Rev. Cytol. 2000, 192, 331–343. [Google Scholar] [PubMed]
- Walter, H.; Brooks, D.E.; Fisher, D. Partitioning in Aqueous Two-Phase Systems: Theory, Methods, Uses, and Applications to Biotechnology; Academic Press: Orlando, FL, USA, 1985. [Google Scholar]
- Albertsson, P.A. Partition of Cell Particles and Macromolecules; Wiley-Interscience: New York, NY, USA, 1986. [Google Scholar]
- Tolstoguzov, V.B. Some physico-chemical aspects of protein processing into foodstuffs. Food Hydrocoll. 1988, 2, 339–370. [Google Scholar] [CrossRef]
- Johansson, H.O.; Brooks, D.E.; Haynes, C.A. Macromolecular crowding and its consequences. Int. Rev. Cytol. 2000, 192, 155–170. [Google Scholar] [PubMed]
- Hancock, R. The crowded nucleus. Int. Rev. Cell Mol. Biol. 2014, 307, 15–26. [Google Scholar] [PubMed]
- Aumiller, W.M., Jr.; Davis, B.W.; Keating, C.D. Phase separation as a possible means of nuclear compartmentalization. Int. Rev. Cell Mol. Biol. 2014, 307, 109–149. [Google Scholar] [PubMed]
- Hancock, R. A role for macromolecular crowding effects in the assembly and function of compartments in the nucleus. J. Struct. Biol. 2004, 146, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Murphy, L.D.; Zimmerman, S.B. Macromolecular crowding effects on the interaction of DNA with escherichia coli DNA-binding proteins: A model for bacterial nucleoid stabilization. Biochim. Biophys. Acta 1994, 1219, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.B.; Murphy, L.D. Macromolecular crowding and the mandatory condensation of DNA in bacteria. FEBS Lett. 1996, 390, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Murphy, L.D.; Zimmerman, S.B. Stabilization of compact spermidine nucleoids from escherichia coli under crowded conditions: Implications for in vivo nucleoid structure. J. Struct. Biol. 1997, 119, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.B. Cooperative transitions of isolated Escherichia coli nucleoids: Implications for the nucleoid as a cellular phase. J. Struct. Biol. 2006, 153, 160–175. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R. Structure of metaphase chromosomes: A role for effects of macromolecular crowding. PLoS One 2011, 7, e36045. [Google Scholar] [CrossRef]
- Zeiger, A.S.; Loe, F.C.; Li, R.; Raghunath, M.; van Vliet, K.J. Macromolecular crowding directs extracellular matrix organization and mesenchymal stem cell behavior. PLoS One 2012, 7, e37904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelly, J.W. Alternative conformations of amyloidogenic proteins govern their behavior. Curr. Opin. Struct. Biol. 1996, 6, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Fink, A.L. Conformational constraints for amyloid fibrillation: The importance of being unfolded. Biochim. Biophys. Acta 2004, 1698, 131–153. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.W. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr. Opin. Struct. Biol. 1998, 8, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Fink, A.L. Protein aggregation: Folding aggregates, inclusion bodies and amyloid. Fold. Des. 1998, 3, R9–R23. [Google Scholar] [CrossRef] [PubMed]
- Bellotti, V.; Mangione, P.; Stoppini, M. Biological activity and pathological implications of misfolded proteins. Cell Mol. Life Sci. 1999, 55, 977–991. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Talapatra, A.; Gillespie, J.R.; Fink, A.L. Protein deposits as the molecular basis of amyloidosis. I. Systemic amyloidoses. Med. Sci. Monit. 1999, 5, 1001–1012. [Google Scholar]
- Uversky, V.N.; Talapatra, A.; Gillespie, J.R.; Fink, A.L. Protein deposits as the molecular basis of amyloidosis. II. Localized amyloidosis and neurodegenerative disordres. Med. Sci. Monit. 1999, 5, 1238–1254. [Google Scholar]
- Lansbury, P.T., Jr. Evolution of amyloid: What normal protein folding may tell us about fibrillogenesis and disease. Proc. Natl. Acad. Sci. USA 1999, 96, 3342–3344. [Google Scholar] [CrossRef] [PubMed]
- Rochet, J.C.; Lansbury, P.T., Jr. Amyloid fibrillogenesis: Themes and variations. Curr. Opin. Struct. Biol. 2000, 10, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Dobson, C.M. The structural basis of protein folding and its links with human disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001, 356, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Zerovnik, E. Amyloid-fibril formation. Proposed mechanisms and relevance to conformational disease. Eur. J. Biochem. 2002, 269, 3362–3371. [Google Scholar] [CrossRef] [PubMed]
- Ferrone, F. Analysis of protein aggregation kinetics. Methods Enzymol. 1999, 309, 256–274. [Google Scholar] [PubMed]
- Dobson, C.M. Protein misfolding, evolution and disease. Trends Biochem. Sci. 1999, 24, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Maroteaux, L.; Campanelli, J.T.; Scheller, R.H. Synuclein: A neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J. Neurosci. 1988, 8, 2804–2815. [Google Scholar] [PubMed]
- Jakes, R.; Spillantini, M.G.; Goedert, M. Identification of two distinct synucleins from human brain. FEBS Lett. 1994, 345, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Iwai, A.; Masliah, E.; Yoshimoto, M.; Ge, N.; Flanagan, L.; de Silva, H.A.; Kittel, A.; Saitoh, T. The precursor protein of non-A beta component of alzheimerʼs disease amyloid is a presynaptic protein of the central nervous system. Neuron 1995, 14, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Weinreb, P.H.; Zhen, W.; Poon, A.W.; Conway, K.A.; Lansbury, P.T., Jr. NACP, a protein implicated in alzheimerʼs disease and learning, is natively unfolded. Biochemistry 1996, 35, 13709–13715. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Li, J.; Fink, A.L. Evidence for a partially folded intermediate in alpha-synuclein fibril formation. J. Biol. Chem. 2001, 276, 10737–10744. [Google Scholar] [CrossRef] [PubMed]
- Eliezer, D.; Kutluay, E.; Bussell, R., Jr.; Browne, G. Conformational properties of alpha-synuclein in its free and lipid-associated states. J. Mol. Biol. 2001, 307, 1061–1073. [Google Scholar] [CrossRef] [PubMed]
- Spillantini, M.G.; Crowther, R.A.; Jakes, R.; Hasegawa, M.; Goedert, M. Alpha-synuclein in filamentous inclusions of lewy bodies from parkinsonʼs disease and dementia with lewy bodies. Proc. Natl. Acad. Sci. USA 1998, 95, 6469–6473. [Google Scholar] [CrossRef] [PubMed]
- Spillantini, M.G.; Goedert, M. The alpha-synucleinopathies: Parkinsonʼs disease, dementia with lewy bodies, and multiple system atrophy. Ann. N. Y. Acad. Sci. 2000, 920, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Tofaris, G.K.; Spillantini, M.G. Alpha-synuclein dysfunction in lewy body diseases. Mov. Disord. 2005, 20, S37–S44. [Google Scholar] [CrossRef] [PubMed]
- Trojanowski, J.Q.; Goedert, M.; Iwatsubo, T.; Lee, V.M. Fatal attractions: Abnormal protein aggregation and neuron death in parkinsonʼs disease and lewy body dementia. Cell Death Differ. 1998, 5, 832–837. [Google Scholar] [CrossRef] [PubMed]
- Galvin, J.E.; Lee, V.M.; Schmidt, M.L.; Tu, P.H.; Iwatsubo, T.; Trojanowski, J.Q. Pathobiology of the lewy body. Adv. Neurol. 1999, 80, 313–324. [Google Scholar] [PubMed]
- Goedert, M. Alpha-synuclein and neurodegenerative diseases. Nat. Rev. Neurosci. 2001, 2, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M. Parkinsonʼs disease and other alpha-synucleinopathies. Clin. Chem. Lab. Med. 2001, 39, 308–312. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Neuropathology, biochemistry, and biophysics of alpha-synuclein aggregation. J. Neurochem. 2007, 103, 17–37. [Google Scholar] [PubMed]
- Uversky, V.N.; Fink, A.L. Biophysical properties of human alpha-synuclein and its role in parkinsonʼs disease. In Recent Research Developments in Proteins; Pandalai, S.G., Ed.; Transworld Research Network: Kerala, India, 2002; pp. 153–186. [Google Scholar]
- Uversky, V.N. A protein-chameleon: Conformational plasticity of alpha-synuclein, a disordered protein involved in neurodegenerative disorders. J. Biomol. Struct. Dyn. 2003, 21, 211–234. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Li, J.; Fink, A.L. Metal-triggered structural transformations, aggregation, and fibrillation of human alpha-synuclein. A possible molecular NK between Parkinsonʼs disease and heavy metal exposure. J. Biol. Chem. 2001, 276, 44284–44296. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Li, J.; Bower, K.; Fink, A.L. Synergistic effects of pesticides and metals on the fibrillation of alpha-synuclein: Implications for parkinsonʼs disease. Neurotoxicology 2002, 23, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Li, J.; Fink, A.L. Pesticides directly accelerate the rate of alpha-synuclein fibril formation: A possible factor in parkinsonʼs disease. FEBS Lett. 2001, 500, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Manning-Bog, A.B.; McCormack, A.L.; Li, J.; Uversky, V.N.; Fink, A.L.; di Monte, D.A. The herbicide paraquat causes up-regulation and aggregation of alpha-synuclein in mice: Paraquat and alpha-synuclein. J. Biol. Chem. 2002, 277, 1641–1644. [Google Scholar] [CrossRef] [PubMed]
- Cohlberg, J.A.; Li, J.; Uversky, V.N.; Fink, A.L. Heparin and other glycosaminoglycans stimulate the formation of amyloid fibrils from alpha-synuclein in vitro. Biochemistry 2002, 41, 1502–1511. [Google Scholar] [CrossRef] [PubMed]
- Goers, J.; Manning-Bog, A.B.; McCormack, A.L.; Millett, I.S.; Doniach, S.; di Monte, D.A.; Uversky, V.N.; Fink, A.L. Nuclear localization of alpha-synuclein and its interaction with histones. Biochemistry 2003, 42, 8465–8471. [Google Scholar] [CrossRef] [PubMed]
- Goers, J.; Uversky, V.N.; Fink, A.L. Polycation-induced oligomerization and accelerated fibrillation of human alpha-synuclein in vitro. Protein Sci. 2003, 12, 702–707. [Google Scholar] [CrossRef] [PubMed]
- Munishkina, L.A.; Phelan, C.; Uversky, V.N.; Fink, A.L. Conformational behavior and aggregation of alpha-synuclein in organic solvents: Modeling the effects of membranes. Biochemistry 2003, 42, 2720–2730. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Li, J.; Fink, A.L. Trimethylamine-N-oxide-induced folding of alpha-synuclein. FEBS Lett. 2001, 509, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Munishkina, L.A.; Ahmad, A.; Fink, A.L.; Uversky, V.N. Guiding protein aggregation with macromolecular crowding. Biochemistry 2008, 47, 8993–9006. [Google Scholar] [CrossRef] [PubMed]
- Munishkina, L.A.; Fink, A.L.; Uversky, V.N. Concerted action of metals and macromolecular crowding on the fibrillation of alpha-synuclein. Protein Peptide Lett. 2008, 15, 1079–1085. [Google Scholar] [CrossRef]
- Munishkina, L.A.; Fink, A.L.; Uversky, V.N. Accelerated fibrillation of alpha-synuclein induced by the combined action of macromolecular crowding and factors inducing partial folding. Curr. Alzheimer Res. 2009, 6, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Xie, J.; Wei, L.; Li, W. Macromolecular crowding modulates the kinetics and morphology of amyloid self-assembly by beta-lactoglobulin. Int. J. Biol. Macromol. 2013, 53, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Fan, J.B.; Zhu, H.L.; Shewmaker, F.; Yan, X.; Chen, X.; Chen, J.; Xiao, G.F.; Guo, L.; Liang, Y. Crowded cell-like environment accelerates the nucleation step of amyloidogenic protein misfolding. J. Biol. Chem. 2009, 284, 30148–30158. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.J.; Minton, A.P. Protein aggregation in crowded environments. Biol. Chem. 2006, 387, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Gaharwar, B.; Gour, S.; Kaushik, V.; Gupta, N.; Kumar, V.; Hause, G.; Yadav, J.K. Assessment of the effect of macromolecular crowding on aggregation behaviour of a model amyloidogenic peptide. Protein Peptide Lett. 2015, 22, 87–93. [Google Scholar] [CrossRef]
- Seeliger, J.; Werkmuller, A.; Winter, R. Macromolecular crowding as a suppressor of human iapp fibril formation and cytotoxicity. PLoS One 2013, 8, e69652. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Fan, J.B.; Zhou, Z.; Zhou, B.R.; Meng, S.R.; Hu, J.Y.; Chen, J.; Liang, Y. The contrasting effect of macromolecular crowding on amyloid fibril formation. PLoS One 2012, 7, e36288. [Google Scholar] [CrossRef] [PubMed]
- Mittal, S.; Singh, L.R. Macromolecular crowding decelerates aggregation of a beta-rich protein, bovine carbonic anhydrase: A case study. J. Biochem. 2014, 156, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Derham, B.K.; Harding, J.J. The effect of the presence of globular proteins and elongated polymers on enzyme activity. Biochim. Biophys. Acta 2006, 1764, 1000–1006. [Google Scholar] [CrossRef] [PubMed]
- Harrison, B.; Zimmerman, S.B. Stabilization of t4 polynucleotide kinase by macromolecular crowding. Nucleic Acids Res. 1986, 14, 1863–1870. [Google Scholar] [CrossRef] [PubMed]
- Harrison, B.; Zimmerman, S.B. T4 polynucleotide kinase: Macromolecular crowding increases the efficiency of reaction at DNA termini. Anal. Biochem. 1986, 158, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Moran-Zorzano, M.T.; Viale, A.M.; Munoz, F.J.; Alonso-Casajus, N.; Eydallin, G.G.; Zugasti, B.; Baroja-Fernandez, E.; Pozueta-Romero, J. Escherichia coli aspp activity is enhanced by macromolecular crowding and by both glucose-1,6-bisphosphate and nucleotide-sugars. FEBS Lett. 2007, 581, 1035–1040. [Google Scholar] [CrossRef] [PubMed]
- Buczek, O.; Green, B.R.; Bulaj, G. Albumin is a redox-active crowding agent that promotes oxidative folding of cysteine-rich peptides. Biopolymers 2007, 88, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.L.; Wu, L.J.; Chen, J.; Liang, Y. Effects of macromolecular crowding on the structural stability of human alpha-lactalbumin. Acta Biochim. Biophys. Sin. 2012, 44, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.; Kundu, J.; Mukherjee, S.K.; Chowdhury, P.K. Myoglobin unfolding in crowding and confinement. J. Phys. Chem. B 2012, 116, 12895–12904. [Google Scholar] [CrossRef] [PubMed]
- Breydo, L.; Reddy, K.D.; Piai, A.; Felli, I.C.; Pierattelli, R.; Uversky, V.N. The crowd youʼre in with: Effects of different types of crowding agents on protein aggregation. Biochim. Biophys. Acta 2014, 1844, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Flory, P.J. Principles of Polymer Chemistry; Cornell University Press: Ithaca, NY, USA, 1953; p. 672. [Google Scholar]
- Goodell, D.J.; Eliseeva, T.A.; Coultrap, S.J.; Bayer, K.U. Camkii binding to glun2b is differentially affected by macromolecular crowding reagents. PLoS One 2014, 9, e96522. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Zhou, Z.; Mo, Z.Y.; Shi, J.Z.; Chen, J.; Liang, Y. Mixed macromolecular crowding accelerates the refolding of rabbit muscle creatine kinase: Implications for protein folding in physiological environments. J. Mol. Biol. 2006, 364, 469–482. [Google Scholar] [CrossRef] [PubMed]
- Batra, J.; Xu, K.; Zhou, H.X. Nonadditive effects of mixed crowding on protein stability. Proteins 2009, 77, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.Q.; Liu, H.J.; Li, C.; Luan, Y.S.; Yang, J.M.; Wang, Y.L. Effects of macromolecular crowding on refolding of recombinant human brain-type creatine kinase. Int. J. Biol. Macromol. 2012, 51, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.R.; Zhou, Z.; Hu, Q.L.; Chen, J.; Liang, Y. Mixed macromolecular crowding inhibits amyloid formation of hen egg white lysozyme. Biochim. Biophys. Acta 2008, 1784, 472–480. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuznetsova, I.M.; Turoverov, K.K.; Uversky, V.N. What Macromolecular Crowding Can Do to a Protein. Int. J. Mol. Sci. 2014, 15, 23090-23140. https://doi.org/10.3390/ijms151223090
Kuznetsova IM, Turoverov KK, Uversky VN. What Macromolecular Crowding Can Do to a Protein. International Journal of Molecular Sciences. 2014; 15(12):23090-23140. https://doi.org/10.3390/ijms151223090
Chicago/Turabian StyleKuznetsova, Irina M., Konstantin K. Turoverov, and Vladimir N. Uversky. 2014. "What Macromolecular Crowding Can Do to a Protein" International Journal of Molecular Sciences 15, no. 12: 23090-23140. https://doi.org/10.3390/ijms151223090